Crossing Forensics Borders is a global lecture series from 14 academic expertise centres in forensic science and education.
Crossing Forensic Borders series

Sessions
Plenary session
Opening session for the Crossing Forensic Borders event, hosted by Dr Scott Chadwick and Distinguished Professor Claude Roux.

Crossing Forensic Borders
Plenary Session
Hello, everybody, and welcome to the Fifth Crossing Borders event today, hosted by the Centre for Forensic Science at the University of Technology, Sydney. I will be your host, Doctor Scott Chadwick. And before we begin today's proceedings, I would like to acknowledge the Gadigal people of the Eora Nation upon whose ancestral land our city campus now stands. I would also like to pay respect to the elders, both past and present, acknowledging them as the traditional custodians of knowledge for this land. So just before we get started in today's session, just got a bit of housekeeping. So with this being an online event, please do bear with us. If there are any technical issues, we will work as quickly as we can to resolve them for you. If you find you're not able to access a talk at any point, it's probably a good idea to maybe log out and then re-log back in, as that would usually resolve some of those issues. If you do have any questions in today's webinar, please do type them in the question and answer box at the bottom of your screen. We will do our best to answer those questions at the end of the session. If you do see a question that you would like to be asked, there is a little up-voting tool at the at the bottom, or right next to that particular question.
Just also letting you know that today's session will be recorded, but we are not recording any video or audio input from the audience, so if you do not wish to be involved or be recorded at any part of this webinar, you may contact UTS at Science.Future@UTS.edu.au. So it's that email address down the bottom there and you can discuss any concerns or questions you may have. Just to let you know as well, after the introduction today, we will open up the parallel sessions and these will be run like a regular Zoom meeting. So when you do join those parallel sessions, please make sure that your camera and microphone are switched off. There will be a final wrap up session after those parallel sessions. So please re-join the meeting that you are in now for any final questions you may have about the presentations.
So to get us started, we are going to have a presentation from Distinguished Professor Claude Roux. So if ever there was anybody that needed no introduction, it would be Claude, but we will give him the introduction he deserves. So, after completing his undergraduate and PhD studies in forensic science at the University of Lausanne, both migrated to Australia in 1996. Claude has been pivotal to the development of forensic science in Australia over the past twenty five years by developing and leading the first undergraduate degree and PhD program in Forensic Science at UTS. He is currently a Distinguished Professor of Forensic Science and the Director of the Centre for Forensic Science at UTS. His Research Activities covers a broad spectrum of forensic science, including microtraces, documents, fingerprints, forensic intelligence and contribution to forensic science to policing and security. Throughout his career, Claude has published more than one hundred and ninety refereed papers, twenty six book chapters and a large number of conference presentations. So if anybody has ever gone to a conference, very likely you would have seen Claude's name there. He has attracted over 5.5 million dollars in competitive research grants over the last 10 years and has received more than 20 prizes and awards. He is currently the President of the International Association of Forensic Sciences, a council member of the Australian Academy of Forensic Sciences and a fellow of the Royal Society of New South Wales. And I will let Claude get started with his presentation today. Thank you, Claude.
Thank you. Thank you, Scott, for a kind introduction and welcome, everyone, and so good morning for people in Australia and good afternoon for North America or America. Good evening for people in Europe. And so it's a pleasure to give you an introduction about the Centre for Forensic Science. And I'd like to thank Arian van Asten and Maurice Alders for proposing this fantastic format of similar series. I think it was absolutely great to have that opportunity, especially given the situation with the pandemic.
So I'll give you first an introduction about our centre. First, University of Technology, Sydney. So first we are in Sydney and is a city in the south eastern side of Australia, which is a big island, and you've got the map here and the university itself is a city university. You really are located in the centre of the city. So it's very attractive to students and staff. It's a young university. So it was created in 1988 and it's the highest performing university in Australia under 50 years of age. The research is excellent.
It's rated as world standard or above in the latest report of Excellence in Research for Australia 2018. The forensic science course started in 1994. So, as you can see, the forensic science course really started very early in the history of the university. So it's been on the map of the university for a very long time. It started from an offshoot from the chemistry degree. But I'll get back to that later. The university has 46000 students approximately, and four and a half thousand staff. The graduate employment is around 70 percent in four months after completion. So it is a very good employment rate. And I must say it's always difficult for people to find a job in forensic science. But if you have UTS degree, it's always a good business card. And we've got a lot of arrangement agreements for student exchange.
Now, our centre. Our centre was created on the back of the very successful first forensic science degree, so the forensic science degree started in 1994. We had the first graduates in 1998. And in 2002, the university agreed to create a Centre for Forensic Science. Our vision is that crime reduction, reduction or fear of crime, crime solving and security in general are major objectives for society. And the mission is to provide an advanced, modern and validated body of scientific knowledge to address questions that are fundamental to the concept of national security and public safety, including intelligence, law enforcement and justice.
So immediately you see that we are not only interested in methods, we are not only interested in the biology or the chemistry of forensic, but we are also interested in how this information is used and used not only for courts, but also law enforcement and justice and intelligence. So that's very, very important.
Now, you know, we're only as good as the people we have are. So I thought I would spend some time to present the members of the Centre. And so essentially the academic staff in forensic science. So you've already heard from Scott or Dr. Scott Chadwick, so he is a UTS graduate, did a PhD in the fingerprint area, is the course director for the Bachelor of Forensic Science at UTS and is doing research in fingerprint, in chemical criminalistics, micro traces in the broad term, criminalistics, etc.. We've got Professor Shanlin Fu who is in charge of our drugs and toxicology program. Shanlin came to UTS more than 10 years ago with a very strong industry experience. More recently, we appointed Dr Anjali Gupta, who is our forensic statistician. So Anjali came from the University of Auckland, so she did a PhD with Professor James Curran over there. Because of the pandemic, she is still stuck in New Zealand. But it's the beauty of virtual courses and especially in her area. So she's been teaching and researching with us for almost a year now. Then we've got Dr Phil Maynard. So Phil came to UTS approximately 20 years ago, and he came after a very strong experience with a local forensic science laboratory in chemical criminalistics. And Phil is teaching and research in fire investigation, he's got some interest in crime scene, he is teaching crime scene and also the broad chemical criminalistics area. We've got Professor Dennis McNevin. Dennis is leading our forensic biology program, is professor of forensic genetics. And Dennis has been very, very active in developing the forensic ancestry and forensic genealogy research programs here and consulting in Australia. Then we've got Dr Georgina Meakin. So she joined it's a bit more than a year ago coming from UCL in the UK, so very strong industry experience and academic experience. She's a forensic biologist, but she's really interested in more the criminalistics component or dimension of DNA. So really, how DNA as a trace can be transferred, persistence, so on. So this is very, very important for us. Then we've got Dr Manoranjan Mohanty is one of the latest appointments. Mano, as we call him, comes from the University of Auckland in New Zealand as well. And Mano is in charge of our newly established digital forensic science major and is developing research in the digital forensic area. Dr Marie Morelato. Marie did her undergraduate degree at the University of Lausanne in Switzerland, but came to do a PhD with us in the area of forensic intelligence. She's developed fantastic research in forensic intelligence and especially drug intelligence. And obviously she's teaching in that area and she's teaching as well in Foundation of Forensic Science. We've got Dr Sebastien Moret, so Sebastien is another Lausanne graduate who came after his PhD to UTS, and Sebastien is a leading force in fingerprint research and also in teaching in areas such as forensic imaging and is teaching in some kind of broad classes like Complex Cases trying to put all the different bits of forensic science together. So Sebastien has a very good track record in fingermark detection. I will skip myself because of the kind introduction by Scott. Then we've got Dr Xanthe Spindler. Xanthe has been around a few Australian universities for her undergraduate and post graduate studies. She did a PhD in fingermark detection with a good friend and colleague, Professor Chris Lennard, at the University of Canberra. But towards the end of her PhD she joined UTS. She's the director for the postgraduate courses in Forensic Science, including the honours degree in Forensic Science, and she's a leading researcher in fingermark detection with also interest in micro traces and chemical criminalistics. Then we've got Professor Barbara Stewart. Barbara has been at UTS almost as long as me. Her area of expertise is in polymer science. So she's been involved in broad chemical criminalistics. And in the last 10 years or so, she's had an increased interest in the composition, especially with our AFTER facility. And I'll get back to that in a moment. Then we've got Dr Maiken Ueland, who did a PhD at UTS in the area of forensic taphonomy, and she's the deputy director of AFTER, our body farm. And obviously she's leading research there. She recently had a major Australian Research Council grant to continue research in that area. Then I'm very cheeky because I put Professor James Wallman, and James is a very good friend of mine and we've been collaborating for many, many, many years. And I sound cheeky because James is the newly appointed Dean of Science at UTS. But foremost, James is a forensic entomologist and is definitely continuing as much as he can research in forensic entomology and collaboration with us. Associate Professor Jody Ward, she's the director of the AFTER Facilities, our body farm. She's on a shared appointment with the Australian Federal Police, which is good demonstration of the partnerships we have. And she's also leading a missing persons initiative with the AFP.
And Anna Wood comes from a very strong industry experience at the New South Wales Police Force in crime scene. And she's been developing a lot of our most recent crime scene teaching tools and she's been teaching in crime scene.
So I thought it was a good idea to spend a bit of time on this, now we also have a very good infrastructure. So we've got a crime scene suite. We've got purpose built forensic science laboratories, and we've got the first body farm in inverted comma, called Australian Facility for Taphonomic Experimental Research or AFTER, the first of this kind outside the US that was launched a few years ago. And all these facilities really are a very strong tool to the students, not only for the research, but for integrated research into teaching. It's very important for the students to get access to these facilities very, very early on.
Now, the pandemic created a bit of an issue for face to face classes at some stage, so we are back on campus for prac classes. But last year, for a while, we weren't. So you can imagine teaching crime scene is a bit of a challenge. And, you know, through the great work of Anna Wood and others, we managed to develop a virtual crime scene immersion facility. And here you've got a few examples, just a few photos illustrating how it could work. So essentially, it was to give a real exposure to crime scene in a virtual manner for students who couldn't come to campus.
Now, our approach is really to try to remove the silos and the barriers from the different disciplines in forensic science. And if you study forensic science, you study forensic science foremost as a transversal discipline. And then you may specialise or you have to specialise in different areas. And then you come back for a capstone semester or a year. So here you have the details. So we start with core forensic subjects or subjects we decided that they were absolutely core for whatever role you have in forensic science ,you should be educated in these areas. So it's Principles of Forensic Science, Forensic Imaging, Forensics Statistics, CSI and Criminalistics. Then the students choose their major if they want to be more orientated towards chemistry material, biology, crime scene and the new major digital. And after the specialisation, the students come back to complete other subjects that we consider absolutely core across the board. And these are complex cases, forensic intelligence, forensic research project, and there are four units of electives to give opportunity to students for exchanges or internship.
So the Biology major, as you can see here, I'm not going to read all the details, but essentially we give, we provide the students the fundamental skills that they need if they are going to work as a forensic biologist or as a forensic scientist in a biology laboratory or related area.
Then we've got to do the same thing for the Chemistry major, so, again, the fundamental skills, a lot of scientific thinking, but also the analytical education that the students need. And here essentially it's for people who are going to work, in a forensic drug or toxicology laboratory or in broad chemical criminalistics, micro traces and so on.
Then we've got the Crime Scene Investigation major, described here, the same principle, but this is more for people who would like to work in crime scene or field or first respondent kind of situations. So there is a bit more specialised subjects like major scene investigation and management, homicide investigation and so on. And the new one, the Digital major that we are developing in collaboration with the Faculty of Engineering and IT so it's to provide opportunities for students interested in this emerging field of digital forensic science. So the digital world created a lot of new avenues for traces. And these traces are obviously they are digital in nature. And as we need, you know, some chemistry to analyse traces that are chemical in nature, we need people with digital or IT skills to to treat traces that are digital.
So our approach has been very successful with the students. You can see on this slide the various implementation and changes in the degree. I think the take-home message here is really the students like that approach. And we have around 600 students enrolled across all our programs at UTS. So it's a pretty large load of students. One thing which is very important to us, it's also the PhD students. We have on average, twenty five PhD students with forensic science projects. And I think this is absolutely crucial because, you know, the research we do with the PhD students is essentially the lifeblood of our research and it's how we can really advance knowledge.
Now, here you've got a description of some of our research strengths. It's by no means exhaustive and I apologise if any of them is overlooked, but it's pretty much the ones that have been around for slightly longer, maybe not for digital forensic science, but we need to flag that one. And we have, you know, for some of them like fingerprints, many, many years of very strong research going on. So, again, we are not doing this research in silos. We try to integrate the various areas and create bridges between the different areas as much as we can we try to provide a transversal approach to forensic science.
Very quickly, some statistics. We bring approximately one million dollars per annum on average, 50 papers, and we have very strong engagement impact, always a lot of very active people. Our main partners, we've got plenty of partners around the world, whether they are end users or in academia. And here you've got some examples of our recent engagement in Science in Focus. We did some great presentation through Georgina Meakin about DNA and we had one about the future of forensic science, where we had Dr Sarah Benson, the chief forensic scientist of the AFP, presenting with Dr Xanthe Spindler and Manoranjan Mohanty.
So obviously, really, we would like to celebrate our people, because we really wouldn't be what we are if we didn't have these very enthusiastic young people. So students and staff. And it's really worth to show a bit of celebration here, what we've done in recent years. A lot of that is related to pre-COVID world, as you can see. It's always good to remind us how good it was when we could meet face to face. But a lot of various conferences, international conferences, posters we really try to promote the students and staff participation in various events, our participation with industry.
And this is a big, big group of our Centre at the latest ANZFSS Symposium in Perth. And we like to have fun. So this is very important to us. We really want to have fun. So this is Amber doing wildlife forensics project. And if people can't find a job in forensic science, we always joke maybe they can find a job in the in the movie industry.
So before finishing, I'd like to remind everyone that in 2023 in late November, so hopefully by then all the international borders will be open. We will welcome everyone from around the world for the International Association of Forensic Sciences Symposium that will be held in Sydney. So I'd like to thank you for listening, and I'm not sure if you've got time for a few questions, but otherwise you follow the instruction from Scott and I hope you find the presentation interesting and informative.
Great. Thank you. Thank you for that Claude. So we do have a few minutes to allow for some questions. So remember, if you did want to ask any of the questions, please put them down in the Q&A box. We do only have a few minutes, though, so if you want to put your question in there, when we return after the parallel sessions, we can use that as an opportunity to get to individuals' questions. So use this time to maybe think about some questions that you might have, pop them in the Q&A and then we'll answer them after the parallel sessions. But Claude, I guess, a question that I have while we might wait, what do you think, what is your sort of vision for the next five years of forensic science?
It's a very interesting and challenging question. I guess it depends if you mean forensic science overall or forensic science education and research. I’ll try to give a general answer. I think we are living in a rapidly changing world. And I think what's going on at the moment with a pandemic, we realise that we are using more and more digital tools and we've got digital ... the digital world is really more and more blended with the physical world. And obviously, you know, through our movement, our activities and the presence of people, we leave a lot of different traces. And the good old world, it was obviously, you know, things like fingermarks and DNA. But increasingly we're leaving our digital traces. So people in digital with, say, digital footprint. So all these traces really have to find a way to be recognised, exploited, integrated with a lot of other physical traces. And I think this is a major challenge because it's a new area. And I feel the forensic science world is still struggling to try to integrate that into the regular forensic science framework, if you want. But having said that, it's not only this, obviously, they are still the challenges in terms of trying to exploit the information we have from traces in a more effective way, especially in the early stages of investigation and even intelligence. And I think the communication in court as well is still remaining challenging and it's an area that has to improve as well. So I don't think there is any secret there.
Ok, great. Thank you. Thank you, Claude. So we are now at the time to transition to the parallel sessions. So you can join those parallel sessions by the link in the PDF. But I will just put them into the chart for all attendees so that they can they can click it in order for those sessions to get started, we are going to need to close this particular session off. So we will just leave it open for a couple more seconds/minutes so that people can get the links and the pass codes. And then if you can join us after the parallel sessions, I will get some more of those questions for Claude. So thank you for coming for the introductions and hopefully you will enjoy the parallel sessions starting very soon. Thank you very much.
Welcome back, everybody. So hopefully you enjoyed the presentations and the parallel sessions, so we're going to use this sort of last few minutes to answer any questions that you may have had from Claude's presentation at the start. So if you wanted to pop those questions into the Q&A, we will do our best to answer them. Now, there were some questions that came through earlier, so we'll just answer those first. So the first question that came through was, do we have any international students in the program? So Claude, I don't know if you wanted me to answer that particular question or...?
Yeah, yeah, I think, Scott, you can answer that. As a course director, you have your finger on the pulse.
So, yes, we do have international students in our programs at the Bachelor, the Masters and the PhD level. In terms of the bachelors, we probably only have about five percent of international students. But as we go through to the Masters and to the PhD programs, we see that the enrolment of international students actually increases. So there are opportunities for international students to study at UTS at whichever level that you're wanting to study forensics at. And so, yes, we do. And it can be accommodated quite, quite easily.
The next question is, do we select students for the program? So this question, again, depends on which program that you're applying for. So the bachelor's and the master's by coursework, those are those are done through a university admissions program. So the students would apply through the processes for international students, that our university offers and that information can be found on our website and then there'll be forms and certain standards that you would have to meet before you would be allowed to study with us. Now with the PhD program, you will still need to apply through the university procedures, but it is always good to reach out to potential academics that you may want to be collaborating with to see if there are any possibilities of doing research in in those particular areas. So Claude at the start gave a list of all the academics that are currently doing research in forensic science. So if you wanted to head to our website, you can find out a bit more about what they do and their contact details, should you wish to do a PhD with one of our academics. Anything to add that Claude?
I think it's quite, quite complete.
Ok, so the next question that we have is in the Masters in Amsterdam, we learn about reporting in the Bayesian framework and is used by the Netherlands Forensic Institute. In Lausanne, the Bayesian reporting is also used. What is the common view of this framework in Australia? And do you report likelihood ratios as well? Or do you report in a different manner? So Claude, I'll pass this.
Thank you. This is a very good, very good question. Um, the framework as such is promoted by the National Institute of Forensic Science in Australia. So we've got a peak body for forensic science. It's called the National Institute of Forensic Science. It's actually Australia and New Zealand. They recently, I think last year, or the year before they released guidelines for evaluation and evaluative reporting, which by and large mimic the ENFSI guidelines. So in terms of broad framework, I think there is promotion and an agreement there. In terms of the actual application, especially with likelihood ratios, I think it's very variable and depending on the trace type. So no surprise, DNA would be a trace type where likelihood ratios are used a lot, but all the types of traces it's less common, especially in when we want to go to the numerical values. So I would suggest to you, go to the NIFS website or maybe we can put that in the chat later and you can get the kind of official view of the Australian community.
Great, thank you, Claude. So do we have any other questions, just quickly, you can type them into the chat. Ah yes, perfect. They have approved of your answer there Claude, so thank you. Thank you for that feedback. All right, so in the interest of wanting to keep everyone to time and making sure that we can wrap up, we might leave it there. So thank you for joining us for the crossing forensic borders. Now, as we mentioned at the start, a recording of this particular event will be announced through social media and made available through our website www.forensics.uts.edu.au. And also on there, you can find out more information about what we do here at the Centre for Forensic Science, the courses we offer and the research that we do. Anybody who has seen a UTS presentation in the last four years will have seen that we have the International Association of Forensic Sciences conference happening in Sydney. Now, this will happen on the 20th to the 24th of November in 2023. And I think by then we will all be very excited to be able to meet again in person.
So if you are wanting to plan ahead, you can book your holidays to come down to Sydney. There is the website, the Facebook page. We also have a Twitter page as well. So you can keep up to date with all the information there. Now, you may have noticed in today's presentation that there were a lot of Swiss accents, and that's no fault, it's not by accident because the Centre for Forensic Science and the University of Lausanne have had a strong relationship over the last twenty five years, which has resulted in student exchange, research collaborations, grant successes, PhD supervision and more than a couple of beers shared over conferences. So it is with great pleasure that we are able to announce that the next event will be on the 3rd of March and it will be hosted by the University of Lausanne. So we hope to see everybody there because no doubt it will be a very exciting presentation. So this concludes the event today. Thank you all for coming. And again, if you do have any further questions, please do get in contact with us. So thank you for joining.
Thank you, everyone.
Fingerprint parallel session
Crossing Forensic Borders Fingerprint Parallel Session, co-hosted by Dr Xanthe Spindler and Dr Sebastien Moret and featuring speakers Teneil Hanna, Romain Steiner and Tim Lee.
Crossing Forensic Borders
Fingerprint Parallel Session
So welcome to the fingerprint parallel session. I'm Dr Xanthe Spindler and the co-host chair. With me is Dr Sebastien Moret. And we're senior lecturers here at CFS UTS and primary supervisors in UTS WSU Fingerprint Group. We're joined by three fantastic speakers that are showcasing a range of research topics in our research group today. We'll save questions until all of the presenters are finished. If your memory is anything like mine, jot down any questions you have so that they're ready to go for the Q&A session. And during the presentations, just a reminder, please make sure your microphones are set to mute. And then once we get to the Q&A session, you can then talk to the presenters. So our first speaker today is Teneil Hanna. Teneil was part of the first Bachelor of Forensic Science cohort, finishing her degree in 2019. She first joined our research group as a third year research intern, expanding on a project investigating the success rates of indanedione zinc and cyanoacrylate fuming. The 14 K Project as it became known for those of you who you may be familiar with that work. Teneil returned to our group last year an Honours student. Her research, which she's presenting today, was focused on fingermark detection and the protocols currently used to assess subsequent fingermark quality. Teneil has very recently, as in the last month, enrolled in the PhD program to expand on her on this research. So, Teneil, if you're ready to go, I'll now hand over to you.
Thank you Xanthe. You'll have to let me share my screen Xanthe. Ok, so just to reiterate, I'm still here and I'm going to be talking about fingermark detection and the best approach to assess the quality of a technique. So just as a basic introduction, fingermark research into new methods of detection and enhancement has continued to grow in decades. However, despite this, there's still a wide range of assessment protocols that are used to assess the quality of these developed fingermarks. Whilst there have been attempts by the International Fingerprint Research Group to standardise assessment techniques, there's still no currently agreed upon approach. So before we even consider the assessments and the scales themselves, we have to consider what is fingermark quality. So to determine fingermark quality, there are several aspects of a fingermark that determine. So first of all, we have things like ridge debridges. Then we have ridge flow and continuity. There's also obviously deal with morphology and pattern. Then we have image quality characteristics like contrast., where you see a high level of contrast, that low level of contrast, as well as background development. And these all lead to the final idealisation of comparison purposes.
So then we move on to the thing about quality assessments themselves, which are trying to determine this fingermark quality. We have two distinct fields this is used in. The first is for research and development purposes. And then we have for comparison and identification purposes. Results of this study, we only looked at research and development purposes and this leads into two common categories, that being subjective quality assessments, which are often quantitative methods to determine the performance of the technique. This is often using scales and visual comparison. And then we have objective quality assessments which these often use software or algorithms, and they give specific values of measured quality. There's currently an abundant number of subjective fingermark quality assessments, which are what I looked into in this study. They are often tailored based on study parameters. They usually adapted from each other. And this has created a very poor cross institutional consistency. And it also asks the question of why do we need so many? So currently there's only actually two absolute scales that are endorsed by the International Fingerprint Research Group, or the IFRG for short, the first being the Centre for Applied Science and Technology, or CAST, Scale. And this is based on a zero to four, which focuses on a clarity of ridge detail. And the second is the University of Lausanne scale, so the UNIL scale and the scale is based on descriptions framed around the quality of what is or isn't visible.
So all of these scales come with some level of limitation. They don't all have these limitations, but they're generally said to be quite undefined. They have no standard for evaluation. They are subjective. That's the biggest problem that we see with these, because obviously they are reliant on human observation. That means they're manual, they're time consuming. They take a while. They're obviously reliant on a human competently identifying minutiae. And then there's also a big problem where we see often placements of fingermarks of different quality placed in the same score categories. So there's no way to actually differentiate these images once quantitatively evaluated, which then leads on to my specific example here, where both of these fingermarks scored a three from the levels of ridge visibility and different levels of contrast. So, for example, on the left, the image here of the fingermark has quite good ridge visibility, whereas the one on the right has quite poor, obviously, because the contrast is quite poor and there's a bit of smudging and superimposition of the fingermark. But they both on the CAST scale, represent as the same. So this leads to the projects and aims of my project. So they were to generate a deeper understanding of subjective assessment protocols, especially the ones that are quite commonly used within forensic research. This then leads to evaluating the efficiency and effectiveness and their benefits and flaws and then hopefully eventually develop and validate a robust grading scheme for fingermark detection techniques. So 10 scales overall were chosen for analysis. All of these scales were chosen based on their ability and the different kind of quality parameters. And so to achieve the category that they have you see here, they had to explicitly mention the parameter and for its use, the smallest areas were background development and confidence of assessors.
And obviously the most represented were usually the ridge detail and visible ridges. All of these scales have been published. They had to be to be used and they were used in prior studies as per these journals seen here.
So, for example, we here have the CAST scale, which was shown before. This actually led onto an adapted scale of the Harush-Brosh Scale, which focuses a little bit more on the comparison side rather than the ridge detail present. So then eventually this was used. I started with a data set of one hundred and twenty representative fingermarks of various quality. These were porous and non-porous surfaces, but only using basic techniques. So just indanedione zinc and then cyanoacrylate fuming and rhodamine 6G. And so this dataset of one hundred and twenty images was chosen based on the original CAST median scores that were given in the original study for this. And then the one hundred twenty images were pooled and ten scales were used and then eight assessors were chosen based on their level of research capability, how much experience they had. So we had some people that were in experience for a year in research in fingermarks and then some that had many years behind them. Then the assessors used a specific assessment program and this software enabled both the scale and a randomised fingermark image. So you can see that example here. So it's quite a simple scoring interface. The program allowed for the randomisation of images so the assessors touch them or randomise them themselves. Timing of assessment as well as comments could be made from the assessments while completing the analysis. And then this obviously led on to a plethora of data which some statistics were made from that. With these results, they were all sorted into categories which kind of represented their parameters as a whole.
And so, for example, the IFRG scales were grouped together, obviously they're the only ones that are endorsed by the IFRG. Then we have things like the adapted scale. So, for example, the Rajan scale which was adapted from a field house study which was adopted originally from the CAST scale. Then we have the non-numerical scales. These ones are the ones that use symbols or letters or words to represent rather than numbers. And then we had the multiple score scales, so these ones represented the scales that had more than one score per fingermark.
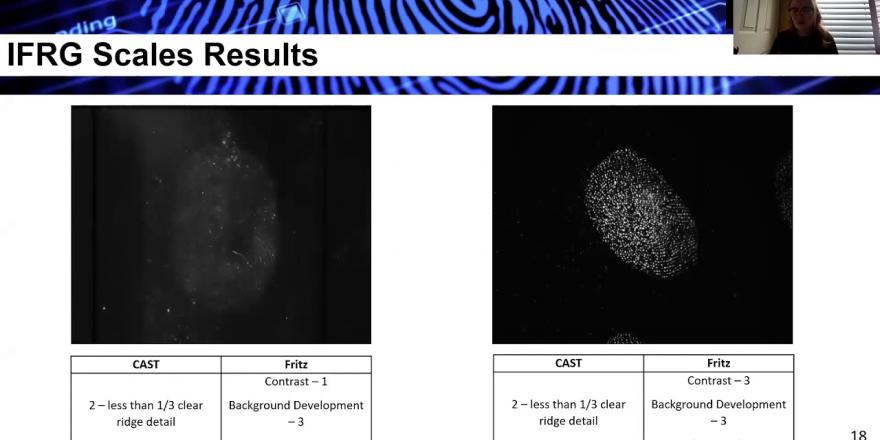
So the adapted and nonnumerical scales indicated the highest level of variability than any other scale, this could have occurred to kind of high levels of subjective interpretation which stems from minimal or too specific descriptions. It's suggested that assessors are less experienced and they are more likely to follow scale descriptors, which was indicated in the study. So finding the ideal symmetry between a minimal description and ones that are too specific to a study or too specific to a fingermark is crucial for understanding how to minimise this variability. The graph here is representative of this seen within these scales. This comes actually, particularly from the RAJAN scale, when the less agreement is noted between assessors for all scores excluding its row. So it was actually anticipated that the scores over zero would be easier for assessors to agree upon, as this kind of makes up the extremes of quality where there's no mark of, or there's mark of the highest quality. So obviously it's quite easy to tell if there's no fingermark at all. Anyone can say there's nothing there, or anyone can say that this fingermark looks perfect. So these marks in comparison to the mixed scores, so the scores that tend to be from a one, two or three, are easier to assess and therefore provide more agreement. This can be specifically noted where agreements in the scores over two or three have a medium agreement of less than 60 percent. So these scales were actually excluded as they showed poor improved protocols, and they were often quite specific to their study.
The multiple score scale results, they showed a little bit more of a larger score range because they had more than once which this inevitably introduced different descriptions to the previous scales. So the additional parameters introduced either too specific or not enough specificity in description, and consequently the scales became more variable, specifically when other quality parameters that assessors weren't used to it introduced. So, for example, here you see the background development from the scale it actually held the highest variability of any scale within this study. These can be attributed to maybe less honest scores, specifically when scoring alongside ridge characteristics in a separate scale. So here we see that there's actually a median score agreement of less than 50 percent in score of one and there's even a zero percent agreement.
And then this could be attributed to the fact that these scales indicate parameters not currently represented in research. So, for example, here we see an image where there is quite poor contrast, but there's very high level of ridge visibility. The CAST scale only gives the score of a two, but the Fritz scale manages to indicate a little bit more into why the ridge detail visibility is so poor. So these results have some impact on the IFRG scale results where issues arise where scores given to two or more different fingermarks, especially middle scores, to nominate both these marks the same. And there's no way to actually differentiate these when you look at a CAST score of a two and a CAST score of a two. So especially once the quantitatively evaluated and popped into graphs. However, the Fritz scale gives a little bit more of an indication here about where it's kind of differing. So contrast is quite poor in one, and contrast is quite good in the other. And the ridge detail is different in both.
So the previous scales were found to have quite specific issues with scale descriptions, subjective interpretation, and they underrepresented quality parameters. And so to further develop the conclusions made in the previous scale, a new robust protocol to assess fingermark was established. So we found, I found that two specific quality categories were found present within these scales. So image quality and fingermark quality and these can be utilised to hopefully differentiate quality in different fingermarks.
So image quality categories are those found from detection techniques. And they're also quite good indicators of how well a technique is working, minus any of the specific kind of mark quality factors. So these include background development and contrast, as seen here. And then fingermark quality categories are those found especially within the fingermarks. So that's things like physical ridges and ridge detail. These are the two common most things found in every scale and all of four of these kind of categories were used within this new scale.
So here is the image quality categories, but now into a new specific description. So each scale was developed using specific descriptors to demonstrate levels of quality found in fingermarks. Specific attention was noted in mid-level scores where variability is quite high. The image quality categories were actually used with words, so qualitatively, and moved into numbers when they were evaluated. This was seen as a... Hopefully would help assessors who aren't used to using background development in contrast scores. And then these are the mark quality categories. So this included adding descriptive levels of amount of ridges present, general morphology of mark and pattern, and there was quite specific reason, ridge detail solely looked at the ridges itself. So obviously, if there's things like level three ridge detail, whereas visible ridges only focused on what you could see, there was no level of detail within this scale. So the results of all of these were piled against the one hundred and twenty fingermarks that were used in the previous assessments for the other scales, and this scale showed middle level scores to have the highest variability, which closely follows that of the previous results for the extremes of quality, as we talked about before, are more easily recognised and agreed upon by assessors.
So background development here, as in the previous scales as well, have the highest variance of any category. And it actually also included a zero percent agreement outlined here as well. This outlier's specifically difficult as it may determine that the median is potentially not the true score that's given by assessors. But this is beyond the scope of this study and will be looked into further. It could be suggested that the lack of experience from assessors has in turn indicated less agreement, especially where the multiple score scales also saw less agreement in categories that did not involve ridge characteristics. However it should be noted that all four categories have results, had a median agreement above 50 percent for every score. So overall, the mark quality parameters had less variability in comparison to the image quality parameters. This category was actually negatively surveyed by all of the assessors, with most stating that image quality parameters are quite difficult to distinguish intechniques that heavily react with background substrates. Which is of particular interest here, because obviously we're using techniques that heavily interact with the substrates.
So a further look into the fingermarks themselves, not just the scale used. Notice quite a big difference in homogeneity and this seems to be a particular pattern that we see in every single scale. These marks tended to sit within the middle scores, which could indicate why current scales show minimal agreement within these marks. So as seen in the example here on both fingermarks. While contrasting background development are not actually different between both of these marks, there's a significant difference noted in the amount of ridges present. So specifically, when analysing the first mark here on the left where the poor contrast areas have diminished some level of ridge visibility and then some areas have enhanced it. This leads to the question of how homogenous they can be considered when assessing quality. It could actually ultimately rely on simply if a technique is homogenous or not. So we could have a simple score to identify if the technique has created homogenous marks. And this could maybe alleviate some of that pressure from the scale itself.
As with all research, the implementation of any new protocol comes with some big limitations. So the scale in itself introduced some level of subjective interpretation. This could be because the assessors had less experience, they've never used this scale before. But subjectivity in itself is a very complex factor. It's well known that subjectivity cannot be completely controlled. This scale did show considerable agreement in its mark quality categories. However, the lack of experience from assessors has led to less agreement in the image quality categories. Ultimately, this could actually lead to the question of how influential is the assessor themselves? And so possibly understanding the level of experience and training required and how assessors determine quality may lead to a less subjective result.
Similarly, this study did not actually use any objective measures to assess fingermark quality. Although it's not within the scope of the study, it should actually be mentioned here that objective methods may pose some benefit in quality assessment. And so this is especially true when considering the results of contrast background development performed quite poorly and these contrast and background development seem to be a big thing in objective means. So perhaps these parameters may be better suited to objective as they could minimise the issue of homogeneity seen.
So overall, in summary, during this project, current protocols of quality assessment have been studied and they showed substantial levels of variability between all of them. The adapted and nonnumerical scales were found to have the highest influence from external factors. These scales may not be robust enough for further evaluation purposes beyond their original studies. The IFRG scale appear to be the most robust of all, which is good because they're endorsed by the IFRG, but they still show issues with some external factors, such as they diminish important quality factors. The Multiple Score scales, while trying to improve on this and introduce these factors missing, but they fail to kind of incorporate them in a way to like increase the assessor agreement. The new scale itself was found to have similar variability to the IFRG and the Multiple Score scales with some improvement on the introduction of quality parameters.
However, there's still limitations. The issue of homogeneity needing to be addressed in order to better differentiate these levels of fingermark quality. So overall, the new scale has made some improvement. But further research is needed to provide a method that can incorporate all quality factors and still remain robust. So to further develop the conclusions made here within this study would involve a level of identifying what is quality. So answer that question, especially how assessors see fingermark quality, and this in turn might help identify the impact of assessor subjectivity. Similarly, more development is needed to understand if quality is better suited to objective means. It's still not known which method is best suited as application of objective systems are often noticed as support tools rather than a replacement. As well, objective systems, they often only focus on one area of quality. So things like ridge characteristic metrics or we also have contrast objective means. And so until there's quite an overall assessment, most find subjective means to be a little bit more accurate as a human can identify everything at once. So the study is only just the beginning of understanding fingermark quality assessments. I'd like to acknowledge the UTS Fingerprint Research Group and then all of the assessors who volunteered their time for this study. So if you have any questions, feel free to pop it in the Q&A. Thank you for listening.
Thanks to Teneil. So, our next speaker that we have up this morning is Romain Steiner. So Romain graduated from the School of Criminal Justice of the University of Lausanne in 2016 after having completed a Bachelor and a Master's degree in Forensic Science with great distinction. His interest for the identification field and more particularly for fingermark detection, led him to complete a master's thesis on the effect of water immersion on VMD processed items. If you haven't read this article yet, it is actually quite an interesting read. Armed with this experience Romain successfully applied for a grant at the University of Technology Sydney to work on a subject dedicated to printing of artificial marks. Since September 2017, he's been undertaking a PhD dedicated to printing of artificial latent fingermarks for improved quality assurance and research efficiency. He's also had the opportunity to work in close collaboration with the University of Lausanne, which has also expressed a growing interest in the development of artificial fingermarks. This collaboration has allowed Romain to spend three months in Switzerland to progress his project and present his results. Besides his research, Romain is also working on in collaboration with Forensic Foundations to help develop a new professional proficiency test kit for fingermark detection, enhancement and identification. So over to you Romain.
Good morning. Good afternoon or good evening, depending on where you're watching this from. As I said, my name is Romain Steiner. I'm a PhD candidate at UTS. And today my presentation will be about the main outcomes of my PhD project, which was dedicated to the inkjet printing of artificial latent fingermarks for improved quality assurance and research efficiency. First of all, I want to speak a short bit about think about fingermark viability and the ways we have to control it or to try to control it. Fingermark variabilities are a well-known issue encountered by researchers when developing new detection techniques or improving existing ones. This variability results from two main parameters, the first one being the chemical composition, which can considerably vary between individuals and for the same individual at different times, and the position factors such as the pressure applied or the angle of contact. For example, during deposition the position of the finger. This variability can lead to obvious differences in the quality of the fingermarks that are detected with the various detection techniques, such as here with ninhydrin, for example.
This variability can lead to discrepancies in the results obtained between different laboratories and the need for standardization has been acknowledged, which led to the publication in 2014 of the IFRG guidelines for the assessment of fingermark detection techniques. This is a twenty seven pages document that aims at getting a more consistent and validated comparable research across groups from all over the world or in other words, this is a standardisation of research. By following these guidelines, the assessment of a new detection technique can be carried out from the initial concept through the final phase implementation. The problem is all those precautions tend to make the entire process very labour intensive and time consuming. And the coordination of all the donors required can be quite challenging because even when clear indications about the deposition method are given to the donors, a full control of the quality still cannot be guaranteed. This is why recent first studies have started to report efforts to control those variability factors. The variability in the physical parameters that occur by designing those kind of what is called fingerprint sampler device like this. I won't go into too much detail, but in short, they allow for the pressure, the time and angle of contact, for example, to be controlled either electronically or by the operator with the help of calibrated springs.
Besides, and this is the most important here, artificial secretions have been designed in order to mimic latent fingermark composition. But those simulants are usually appropriate for a few detection techniques only.
Regarding the control of the chemical composition, it is quite obvious that the fingermark composition itself cannot be controlled because it is very variable by nature. Instead, using standard solutions containing some target compounds with a known concentrations seems to be a promising alternative. However, while those standard solutions are practical to produce, for example, test strips or quick assessment of the detection technique performance, they are they are usually limited to a few compounds and cannot be used to assess detection sequences, for example. And for these reasons, it is legitimate to wonder if those standard solutions can be considered as actual fingermark simulants. Many different secretion formulations have been reported in the literature, but in most of the cases, they are limited to synthetic sweat solutions, which are made of a mixture of amino acids and other compounds, such as inorganic salts, are sometimes added as well. There are only a couple of studies that have reported the use of more complex salt solutions in the form of emulsions of eccrine and sebaceous compounds. So this shows that there is still a clear gap in the literature regarding synthetic secretions.
Following the same trend in synthetic solutions, commercial fingermark simulants have started to appear on the market. They are usually found in the form of those kind of pads, which are soaked in the artificial solutions. These pads were already started in 2013 by Zadnik and co-workers. And I also had the occasion to work with those pads for my research. And in both cases, it was shown that even if the use of those pads give a better control of the quality, in some cases they are too unreliable and cannot be recommended for use in research or practice. These observations brought me to the conclusion that we can definitely do better in terms of artificial fingermarks, and this is what this project was all about.
The main aim of the research was to present the method to controllably and reproducibly produce artificial fingermarks. The first half of the project, the first part was dedicated to the design and optimisation of artificial secretions, the best of their reactivity towards a commonly used detection techniques, as well as their compatibility with inkjet printing is can be considered as being the control of the composition, which is the first parameter in fingermark variability. The second part focused on the optimisation of the inkjet method, its reproducibility, as well as a quality assessment of printed fingermarks processed with different techniques which can be considered as being the control of the deposition. We now talk about... So what I did to develop my own artificial secretions. First of all, the synthetic sweat and sebum solutions that I used were based on the work undertaken by Mackenzie de la Hunty for her PhD thesis in 2017. After this the synthetic sweat and sebum solutions were emulsified to create the final artificial secretion. And it was done as follows. So a small quantity of sebum must pipetted in a test tube. The dichloromethane then that was used to dissolve all of this of the synthetic compounds was evaporated to obtain the dry sebum. Synthetic sweat was then added to the dry synthetic sebum with a ratio of sebum to sweat of one to five. And the mixture was then sonicated for 10 minutes and placed in the fridge for 30 to obtain this solution. This is an emulsion that had the appearance and consistency of milk. It should also be noted that I also produced a concentrated emulsion by using a concentrated sebum, so two different emulsions were used in the project. The reactivity of the emulsion was tested towards compatible detection techniques. Indanedione/zinc, ninhydrin, Oil red O and physical developer. And this was done with the use of spot tests deposited on copy and filter paper.
From the results, it can be seen a good reaction was obtained when the spot tests were processed with the two amino acid agents. The slight differences in colouration of the ninhydrin treated spot test were attributed to differences in the chemical composition of the paper. Because actually the same difference was observed when real .. were processed. Regarding the overall results, it can be seen that increasing the sebum concentration in the emulsion related to a stronger staining when processed with OrO as expected, since it is a lipid stain. However, you can see that the fraction of ... the sebum fraction of the emulsion seems to be concentrated in the centre of the spot and does not migrate in the same way as the eccrine fraction, which is one of the problems when spot tests are pipetted on porous substrates, which tend to separate the different compounds. For the physical developer, deposition was observed on the edge of the spot tests, that was deposited on copy paper. But this was not observed on filter paper. And interestingly, no increase in sebum deposition was observed when the concentrated emulsioni was used, with the spots remaining mostly undetected. So those results showed that, with the exception of PD, which is a very complex technique that is still not fully understood nowadays, the emulsions seem to react to the to the detection techniques in a similar way as real fingermarks. Moreover, the artificial secretions were liquid enough to be printed. Which brings us to the next and main part of the project, which is the actual inkjet printing of artificial fingermarks.
Three deposition methods are usually described in the literature to deposit artificial solutions. The pipetting method is quick and efficient, but the solutions tend to migrate in the substrate in a homogeneous way, which leads to unreproducible spot shapes, as was observed, for example, with the OrO on the previous slide.
Because of this, it was deemed as being impractical and was excluded for the rest of the project. The stamping method makes it difficult to control the amount of material that is loaded on the stamp and then deposited on the substrates. And some compounds may also have a particular affinity with the rubber of the stamp. So for this reason, it was also excluded. For the printing method, research published showed a very good reproducibility and the method has the clear big advantage to allow deposition of fully personalised and customisable patterns. And for those reasons, it seems to be the best method to produce artificial fingermarks in a reproducible way.
So this is how the process works. Which is what I did to print my fingermarks in the project. Well, first of all, the emulsion was prepared by following the methodology presented a bit earlier. So whereas synthetic sweat and sebum solutions will combine before the emulsions. After this, black HP cartridges was emptied, thoroughly cleaned and the ink was replaced with the emulsion. The cartridge was placed in a HP deskjet printer, which is a thermal inkjet printer. And fingerprints were printed with the emulsion.
The printed fingermarks all contained a mixture of eccrine and sebaceous compounds and so multiple target compounds for detection techniques. The composition was controllable and the fingermarks obtained were reproducible as no significant differences were observed in the amount of synthetic secretions printed on all the different A4 sheets. And this was based on a test that was conducted on more than four hundred pages printed.
Fingermarks, were printed with different synthetic solutions. So the synthetic sweat and the synthetic sebum solutions. The sweat and the sebum in sequence, so one after the other. And the concentrated emulsion which was chosen over the original one as it was shown to work better with OrO. For the synthetic sebum, tests show that the synthetic sebum dissolved in dichloromethane was drying very quickly in the cartridge and it was therefore difficult to print. And as toxicity was also an issue, it was decided to replace the DCM by a 50:50 hexane:isopropanol mixture, which is less volatile and toxic. The general scheme of the experimentation is presented here. Only the most interesting results will be presented in the following slide. Otherwise, this presentation will last for 40 minutes and not 20.
So here are some of the results. So for the artificial fingermarks printed with the concentrated emulsion and processed with individual detection techniques, all showed a very good quality at the exceptions of the fingermark treated with the physical developer that all remained undetected. Different tests were undertaken with fresh and older artificial fingermarks, but no ridge detail was observed in any of the cases with the physical developer. When artificial fingermarks printed with the same concentrated emulsion were processed with the detection sequence on paper, the ninhydrin reaction was quite faint compared to the reaction observed when it was applied on its own. And this decrease was attributed to a decrease in the amount of amino acids available for the reaction after indanedione/zinc, which is also an amino acid reagent. The age of the emulsion was shown to have a clear impact on the quality of OrO detection because no ridge detail was observed when the 10 week old emulsion was used, and a better quality was obtained when a fresh concentrated emulsion was used. In either case, the OrO results were not as contrasted and defined as always OrO on its own. It might be explained by the use of press for the and the processes that may have an impact on the sebaceous fraction of the emotion. VMD results were interesting as well. The eccrine artificial marks were almost invisible after VMD was applied. Sebaceous artificial fingermarks, vary in intensity after processing with an absence of metal deposition on the ridges in the same way as what is usually observed with real natural fingermarks. Ridge detail was visible, but with very limited resolution. Artificial fingermarks printed with the concentrated emulsion where halfway between the purely eccrine and sebaceous fingermarks in terms of contrast. But their overall quality was superior to what was observed with sebaceous marks.
So a number of observations were made during those experiments. First of all, it is known that using ORO in sequence after the amino acid reagents can have an impact on the quality of the fingermarks due to the use of non polar carrier solvents with the amino acid reagents. The amount of fatty acid emulsion is probably not as important as in real fingermarks due to the form of the emulsion, which starts to be a bit different from real fingermark residues and may be more impacted by prior amino acid reagents. The loss of lipids over time was demonstrated by the loss of reactivity with ORO when the three month old emulsion was used. And the degradation of lipids in the emulsions seem to follow a similar pattern as real fingermarks as none of fingemarks, either real or artificial, showed any reaction after seven days in an oven at 80 degrees. Which means that even when the emulsion was fresh, good results were not always warranted.
Some limitations were highlighted during those experiments, some of them having been mentioned already. First of all, this may be the most important that no traces of sebum deposition were observed for any of the fingermarks treated with the physical developer. The issue was found to stem from the emulsion itself, and it could not be excluded that the amount of material deposited on the substrate with the printing method was not sufficient enough to be detected by the technique or that some target compounds are still missing to trigger sebum deposition. There are some obvious limitations in the choice of the substrate and the types of solutions that can be used. Substrates have to be thin and flexible to be put in the paper tray. So, for example, commonly found substrates such as glass, for instance, cannot be printed on, which is quite an issue. Besides, the solutions must have a low viscosity, which is not ideal on a non-porous substrate, where the solution will tend to just bleed on the surface. The cartridges have to be modified to be used with the synthetic solutions. It takes some time and has to be done properly to ensure good quality. Some issues may also arise regarding the availability of the cartridge in the long term, especially when a commercially available printer is used. So in conclusion, the emulsion used in this study was found to be a good compromise between complexity and reliability because it requires a reasonable amount of compounds while being reactive towards many of the most commonly used detection techniques. Because the emulsion contains eccrine and sebaceous compounds, it allows for detection techniques targeting different compounds to be tested in a one step process and also allows sequential processing. The use of the HP printer allow the production of good quality artificial fingermarks, and the method was shown to be highly reproducible and quick, with more than one thousand fingermarks that can be printed in high quality per day. And one of the current shortcomings is the stability of the emulsion as it was shown that the quality of the results was dropping when an older emulsion was used, mostly because of the loss of lipids over time.
And finally, and this is the last slide, because I saw that I'm running out of time. I just want to give a few words about the potential applications of artificial fingermarks. There are three of them that seem particularly promising. The first one is proficiency testing. Last year, I made a collaboration with a proficiency test providing company where fingermarks were printed on different documents and sent to different laboratories.
The main advantage is that there is a great control of the quality of the fingermarks, and any failure by the laboratories to detect the marks can be attributed to the detection techniques used and not for the quality of the depositions.
And the is the way to control. When natural fingermarks are used for proficiency testing, it cannot be guaranteed that all participants will receive fingermarks of equal qualities. And any failure to detect those marks can be quite challenging to assess due to fingermark variability. This may be the most promising application of artificial fingermarks in practice. More details about those proficiency test can be found in the final report that was published last year. And given the success of this approach, an improved version is already planned this year.
Artificial fingermarks could also simplify cross-laboratory comparisons. Research is a continual process, and when the techniques are developed or improved, having control fingermarks will help towards easier comparisons between the results obtained by different research groups.
Finally, test strips are already being used by some laboratories to assess the performance of their reagents, and test strips bearing artificial fingermarks might be very useful to control the quality of detection technique in the one step process. So that's all for me. Sorry for overstaying a little bit. So if you have any questions, keep them in mind? And we will answer at the end of this session. Thank you very much for your attention.
Thanks Romain. So we'll move on now to our final speaker and I can see questions starting to pop up into the chat. So this is great. We'll have a great Q&A session at the end. So our final speaker is Timothy Lee. Tim is a PhD candidate at Western Sydney University with his PhD thesis currently under examination. He completed our old Bachelor of Forensic Science in Applied Chemistry degree in 2015 and returned the following year for an Honours project comparing single metal deposition and physical developer. Tim developed a strong passion in the discipline of forensic identification during his undergrad and Honours studies and wasted no time in jumping on the opportunity to apply for a competitive Australian Research Council funded PhD scholarship, working on further research into nanoparticle reagents for fingermark detection. This morning, he'll be presenting on his PhD research using functionalised silicon oxide nanoparticles for latent fingermark development. Take it away, Tim.
Thank you Xanthe for that introduction. And hi everyone. My name is Timothy and welcome to our presentation. So. Without further ado, let's get into it. When it comes to fingerprint evidence, it has always been the gold standard of forensic identification, as we all know, and some of the conventional techniques that are available for us to use include powder dusting, amino acid reagents such as indanedione/zinc and ninhydrin. However, there are limitations that we are facing now with some of these conventional techniques. The major limitations are fingermark sensitivity and selectivity. According to a study, certain techniques actually struggle to detect some fingermarks that are present under certain conditions. In terms of fingermark selectivity, methods such as cyanoacrylate fuming and physical developer, the exact components that they are targeting in fingermark residues are still not conclusively determined. So how can we actually deal with these limitations? Luckily, we have nanoparticles. Nanoparticles are something that we can use to help overcome these limitations. There are three distinct characteristics that are desirable for latent fingermark development from nanoparticles. Firstly, the surface of nanoparticles is modifiable, meaning that we can improve fingermark selectivity by utilising different molecules to help establish selective interactions with fingermark residues. Second of all, some nanoparticles can be internally functionalised. For example, the core of nanoparticles can be filled with luminescent dye. And this is obviously advantageous for fingermark development on surfaces with complex background by having luminescent fingermarks. Lastly, the average spherical diameter of most nanoparticles is less than 100 nanometres. Such small size is beneficial to produce developed fingermark ridges with high resolution.
So it sounds so nice and tight so far, right? But as far as research goes, we all know that there's always a catch. Where in this case, the problem is that not all nanoparticles possess all the three characteristics that are desirable for latent fingermark development at the same time. This means that we cannot fully exploit the potential benefit from using nanoparticles for latent fingermark development. I mean, kind of a bummer, right? But what if I told you there's actually no catch? And that's right! The plot twist in this case is called silicone oxide nanoparticles.
So what's so special about silicon oxide nanoparticles? First of all, a range of different molecules can be incorporated with silicone oxide nanoparticles so it can offer high fingermark selectivity. Furthermore, a wide range or wide selection of luminescent dye molecules can be used to modify the optical properties of silicon oxide nanoparticles to produce luminescent fingermarks. Also, homogeneously distributed nanoparticles can be easily synthesised. So a combination of the three characteristics is highly desirable for improved fingermark sensitivity and selectivity. For example, compared to conventional methods such as powder dusting, fingermarks developed using functionalised silicon oxide nanoparticles are luminescent, of course, with much finer fingermark ridge detail.
So in 2016, a preliminary study provided useful insights into using functionalised silicone oxide nanoparticles for latent fingermark detection. In a study, authors concluded that the functionalised silicon oxide nanoparticles that they utilised, which was a carboxyl-functionalised silicon oxide nanoparticle base region was promising to provide improved fingermark selectivity and sensitivity. So let me quickly go through how the method works here. Firstly, the water base RuBpy-doped CES silicon oxide nanoparticles are synthesised. And in terms of the application procedure, fingermarks are first rinsed in water in a water bath before being immersed in a colloidal dispersion which is made up with the nanoparticles, of course, for 60 Minutes for fingermark development. Finally, the processed fingermarks are rinsed in water for five to 10 minutes. And in terms of visualisation, just really similar to any other luminescent fingermark technique, once you can see fingermarks are developed, you just then move on to the imaging step. So although this preliminary method demonstrated that the potential of using functionalised silicon oxide nanoparticles for fingermark detection, optimisation was definitely needed to improve the overall practicality. So based on this, my PhD began with the optimisation of the nanoparticles for fingermark detection. So as I mentioned in the previous slide, or as you may have seen from the animation, the synthesis process of the nanoparticles takes 48 hours to complete and the colloidal dispersion in the original method was made up by diluting the nanoparticles by half. So I wanted to know if I could get away with using a more diluted colloidal dispersion, meaning that I want to see how would fingermark detection quality be affected with different nanoparticle concentrations? Well, surprisingly, it was concluded that a much lower concentration of nanoparticles using the colloidal dispersion is in fact the optimal concentration to develop fingermarks.
Well this is because at high concentrations, the surface functionalised nanoparticles encounter nanoparticle attractions and these attractions prevent effective interactions with fingermark residues. And after that, since the fingermark immersion time in the colloidal dispersion was 60 minutes long at room temperature in the original method. And it was obviously not practical for routine application. And I also looked into reducing the treatment time. In the original study, it was suggested that chemical bonding is the major interaction between nanoparticles and fingermark residues. So from a chemical viewpoint, increasing the temperature of the colloidal = dispersion would increase the reaction rate and allows for a shortened version time. And lo and behold, we found that fingermark detection quality was far better with a higher treatment temperature and a much shorter immersion time was required. Our comparison study between the two sets of detection parameters also showed that the optimised detection parameters gave superior results. So to summarise, in comparison to the preliminary method, it was determined that a 40 times dilution of the nanoparticles is the optimal concentration to be used to make up the dispersion. To achieve a 40 degree fingermark treatment, application was conducted using a hot plate, and in contrast to a 60 minute immersion time, treatment time was reduced to only five minutes because of the higher treatment temperature.
And here are a few example fingermarks on different substrates from the comparison study between the two sets of detection parameters. Note that the left fingermarks were developed using the preliminary method, while the right fingermarks were treated using the optimised method, as we can see here. And it is clear that the fingermark detection effectiveness was superior with use of the optimised detection parameters. So although the optimised method demonstrated better fingermark detection effectiveness, using a hot plate for fingermark treatment, was a bit, to be honest, a bit iffy. So to improve technique, although for technical applicability, a shaking incubator, as the one that we can see on the right hand side here, was incorporated into the treatment process for several reasons. First of all, it provides accurate control of temperature for the treatment process. And the large inner chamber also means that larger and more items can be processed at the same time. For the nanoparticles, optimisation of the carboxyl SiO surface functionalisation was also completed. The amount of CES molecules used for surface functionalisation was not optimised in the original study and in our research it was concluded that 50 percent of the original amount to be the optimal concentration for this particular functionalisation for latent fingermark detection.
So with the latest optimisation, let's now look at the recommended treatment protocol using the nanoparticle based regime for fingermark development. So as we can see here, focusing only on the colloidal dispersion, the nanoparticles are synthesised with the optimal amount of CES, which is, again, the molecule used for carboxyl surface functionalisation, fingermark treatment is done using, with the use of shaking incubator. So to evaluate the fingermark detection effectiveness of the latest optimised method, the two treatment methods were compared once again. Here are some example fingermarks from the experiment on different substrates. So the left fingermarks were fingermarks treated using the original CES with the hotplate treatment method, and the right fingermarks were treated using the optimal CES amount with the use of a shaking incubator. So it was observed that the further optimised parameters provided improved fingermark detection effectiveness on the evaluated fingermarks. From the fingermarks that we can see here, this is particularly more pronounced on the second and third fingermarks. The ones on the two plastic substrates here, transparent polypropylene and green polyethylene.
So with the latest optimisation, comparison of the nanoparticle base method with a benchmark technique to step CF fuming was completed and the results showed that on average, two step cyanoacrylate fuming demonstrated better detection effectiveness on the evaluated fingermarks. And the nanoparticle based method was actually concluded to be less donor dependent and more substrate-dependent. And this could be viewed as an advantageous trait for case scenarios where wide donor variability is expected.
So the example fingermarks here are from the comparison study between the two step, between two step cyanoacrylate fuming and the nanoparticles. They are representative fingermarks on the six test substrates used in the experiments. The left halves were treated using two step cyanoacrylate fuming while the right ones were treated with the nanoparticle based method. As I mentioned in the previous slide, the two step cyanoacrylate fuming performed better on average on the evaluative fingermarks. But what I want to really highlight here is an advantage of using the nanoparticles for fingermark development. So on the aluminium foil and the two plastic substrates here, the development of fingermarks with nanoparticles generated more homogeneous and continuous ridges as we can see here. Other than that, it was also observed that the two techniques were generally less viable for fingermark detection on glass, white gloss paint and white ceramic tile in the study.
So like I said earlier, the nanoparticle based method was less donor dependent and more substrate dependent. Here's some example fingermarks illustrating the detection effectiveness of the two methods on impressions from two weak fingermark donors. It was observed that two step cyanoacrylate fuming was effective on the evaluated fingermark specimens from Donor A.
While it exhibited low fingermark detection effectiveness on impressions from donor B. Even though some kind of inter-donor variability between these two donors was expected, the observed difference in detection effectiveness for two step cyanoacrylate fuming, on impressions from these two was again quite weak fingermark donors was still unexpectedly high. So with generally poor results for fingermarks from donor B. In contrast, the nanoparticle based method was able to generate visible detail in the fingermarks from donor B on the two plastic substrates. Traces of development were also shown on the aluminium foil.
So all right, so moving on here to tests of the compatibility of the nanoparticle based method with routine techniques, it was applied in sequence with two step cyanoacrylate fuming, and the results indicated that the two techniques are not compatible for application in a detection sequence. So if the nanoparticle based method can't be a prior sequence with routine techniques, can it be used in sequence on its own, meaning multiple repeated development of fingermarks using the nanoparticle based method? The results confirm that by repeated application, overall fingermark detection quality increased significantly across the evaluated fingermarks. And here are some example fingermarks from using the nanoparticle based method for multiple sequential treatments. On each fingermark, the last half was treated using the nanoparticle based method once, while the right ones, right half, underwent two additional treatments, except the ones on aluminium foil, which underwent only one additional treatment.
So it was observed that the detection quality was improved using multiple treatments of the nanoparticle based method. There was a slight increase in ridge clarity resulting from the second fingermark treatment and a significant increase in ridge clarity and ridge detail resulted from the third treatment, as we can see on the two plastic substrates here in particular. So up until this point, the nanoparticle based method had only been used for application in a laboratory setting. So was it possible to use it outside of a lab, maybe, let's say, at a crime scene setting? In order to investigate this, the nanoparticle reagent was also applied as a spray. OK, so here are some representative fingermarks treated using the nanoparticle reagent as a spray. As a trial experiment, there was heavy background staining on almost all of the fingermarks. This is most likely due to the insufficient amount of water used for rinsing during the experiment. But the main point that I want to focus here on is traces of fingermark development were able to be produced from the spray treatment across different substrates and donors. As we can see here.
So after that, to assess the ability to subsequently enhance fingermarks detected via this spray application, half of the treated fingermarks in the above experiment underwent treatment using the silicon oxide nanoparticle based method in a colloidal dispersion bath, which is the method that I mentioned earlier, which are the right halves of the fingermarks that we can see here. We can see that both the finger ridge detail and clarity, we improved by the subsequent treatment using the nanoparticle based method in a dispersion bath. However, the subsequent treatment bath was not able to remove or reduce the extent of background staining on the fingermarks resulted from the preceding spray application.
So to give a quick idea, as in how the nanoparticle spray works, here's an illustration. With the nanoparticle based reagent in a trigger spray bottle, let's say, is applied to the surface of interest. The treated surface will be left untouched for one minute to allow for fingermark development. The surface is then sprayed with water to remove background nanoparticle droplets. Finally, the quality of the developing fingermarks can be assessed with the naked eye, for example, using a portable forensic light source. Although it's great to see that the proof of concept experiments show affirmative results for the nanoparticles reagent to be used as a spray, one thing I want to emphasise that more comprehensive assessment will be required for it to be considered for operational implementation. So these are just proof of concept experiments.
So in conclusion, an optimised RuBpy-doped CES silicon oxide nanoparticle based method was proposed for latent fingermark detection and enhancement. It's not compatible with benchmark techniques such as cyanoacrylate fuming now. It can be applied in repeated fashion to increase overall fingermark detection quality. And the possibility of using the optimised nanoparticle based reagent for operational use at crime scenes was demonstrated via spray application, and the benefits of exploiting the nanoparticle based method for latent fingermark development were demonstrated. And I also want to emphasise, we have to continue to explore the advantages of using silicon oxide nanoparticle based method for latent fingermark detection and enhancement.
So before I actually finish, I would like to acknowledge the Australian Research Council for funding this research as a linkage project, and I'd also like to thank ROFIN Australia, the Australian Federal Police and the Victoria Police for their contributions as the partner organisations. So if you have any questions, feel free to jot down any in the Q&A session and you can also contact me by this email. And that's it for me. Thank you.
Thanks, Tim.
Forensic Biology/Taphonomy/Wildlife parallel session
Crossing Forensic Borders Forensic Biology/Taphonomy/Wildlife Parallel Session hosted by Dr Georgina Meakin and Dr Maiken Ueland and featuring speakers Jeremy Watherston, Sharni Collins and Amber Brown.
Crossing Forensic Borders
Forensic Biology Taphonomy and Wildlife Forensic Science Session
Ok, so good morning or afternoon, depending on where you are at the moment, and welcome to the Forensic Biology Taphonomy and Wildlife Forensic Science Session. As with the main session, this session is being recorded. We're also going to leave questions and answers until the end of the session. And for those of you who don't know me, my name's Georgina Meakin. I'm a senior lecturer here at UTS and will be chairing this session. Please bear with us. A lot of people are still entering into the Zoom sessions. We'll be admitting as we go. But in the interest of time, I would like us to move forward, so I would like to go ahead and introduce our speaker. That's Jeremy Watherston. Jeremy is a candidate here at UTS. His research focuses on the recovery of DNA from compromised human remains and DNA-based disaster victim identification. And this research is conducted in collaboration with AFTER. Jeremy's research includes optimal sample selection, collection and preservation, as well as novel profiling approaches and the application of rapid DNA platforms, some of which we will hear now. So Jeremy, over to you, so please share your screen.
Thank you. Thank you, Dr. Meakin. So you see today I'll be talking about optimisation of rapid DNA profiling from compromised human remains.
So first of all, if I could just do a few acknowledgements, this project is supported by an Australian government research training program scholarship, and the research is sponsored by New South Wales Health Pathology Forensic and Analytical Science Service. I'd also just like to give acknowledgments to AFTER and especially the selfless donors who made this work possible.
So the work presented, has been approved by four human research ethics committees as well as for governance.
And if I could just give a quick outline of what I'll actually just go through today, first of all, I'd just like to start with a bit of a background for disaster victim identification before focusing then obviously on DNA based identification. I'll just briefly touch on traditional DVI guidelines. And that's really with regards to optimal sample types, as well as how these samples are prepared for later DNA processing steps. Then we'll have a look at the literature and what the literature has come forward with regards to new sample types, the way that these sample types are prepared, as well as the new technology that's really emerged to allow us to be able to do identification faster, cheaper etcetera, and possibly with more information. Then I'll just go through a couple of my experiments. And that's really within the context of our work on rapid DNA platforms, rapid approaches to DNA processes. And that's within the context of what we've done with preservative solutions, direct PCR methodology within a surface decomposition study at AFTER, rapid DNA platform testing and then burial decomposition study. And then I'll just finish up with a couple of conclusions.
So just as a courtesy, please be advised that there are images of deceased persons in an advanced state of decomposition, and I just ask that, that no photographs or screenshots within the current forum just in respect of our donors.
So following a mass disaster, one of the first priorities is the identification of victims. So Interpol reports three primary identifiers, specifically fingerprints, DNA and dental records, or odontology, and DNA based identification has been reported as the gold standard. That's due to its ability to identify victims and reassociate remains, as well as provide investigative leads. And that's all at a relatively low cost and with a high degree of discrimination. So traditionally, femur and teeth samples have been targeted for DNA based identification with femur samples being reported as the optimal sample type. So that's really because the physical structure of these sample types, as well obviously the bone in the bone matrix really affords DNA within these samples that extra protection.
So whether that's protection against the environment, so we're talking about tropical, hot, humid environments; or whether that may be an insult due to the nature of a disaster, for example. So if we're talking about fire, et cetera.
However, what we can see is that this actually requires, to be able to sample femur requires a laborious, time consuming surgical procedure to remove a femur which from the deceased. So if you look at the pictures, say that an H cut is made to expose the femur and then a bone saw is used to basically cut out that bone wedge. But even then, even once that femur wedge has been sampled, still has to undergo quite a lengthy, extensive, laborious preparation, cleaning, and then even the DNA processing steps itself is quite extensive.
But if we review the literature, we can see that a large number of novel postmortem sample types have really emerged in the last five or so years. So a number of these sample types offer an opportunity for a faster DNA based identification. And that's really due to a simpler method of collection, preparation, processing and the DNA testing steps itself. So these sample, these novel postmortem sample types, or the new sample types, have also been reported to give a good yield of DNA, which is arguably the limiting factor in that step. So and that's even in compromised human remains cases. So that the novel sample types that that I focus on in my research is nails and distal phalanges of the hands and feet.
So combined with a plethora of commercial DNA testing kits that are far more forgiving with regards to inhibited and degraded samples, as well as the multiple emerging DNA technologies, there really is an opportunity for the international DVI standards and guidelines to be revised to reflect current thinking about sample selection, collection, preservation and DNA profiling.
So we started with preservative solutions, so these preservative solutions preserve at room temperature, but they also cause that DNA to leach into solution. Then we sought to apply this preservative solution to an automated direct to PCR workflow. So in that directed PCR approach we're removing the cell lysis extraction and quantification steps, which really reduces the costs as well as the time frames. The literature also reports that it's associated with an increase in sensitivity, which is obviously of benefit. However, because we are removing that extraction step, which effectively cleans and separates out the DNA from an otherwise dirty sample, it's possible that inhibiters could remain. And because it's going straight to the amplification step, the reaction could be susceptible to those inhibitors.
So we tried this approach on multiple 30 year old bloodstains, so I just got a picture here of two replicas of the same stain and what we were able to do is actually obtain better results when compared to existing laboratory standard operating procedures. But we did actually have to refine the solution itself. And that was, as I briefly touched on, ultimately because the preservative solution that we use does contain known inhibitors. And without that extraction step, it was susceptible to problems with the amplification step.
So we then extended trials to blood, saliva, hair and nail samples. So while this testing was associated with faster processing times, lower costs and storage solutions, the DNA recovery wasn't as good in this case. So after further trials, we believe that this may have been due to insufficient cell lysis, particularly in sample types such as the nail.
So thinking about, in the direct PCR methodology, you're really relying on the high temperatures associated with the denaturation step of DNA to facilitate that cell lysis. So we just believe without the existing extraction steps, especially for the sample types such as now, that there may have been issues with actual successful cell lysis.
So next we wanted to test our new sample type, so specifically the nails and distal phalanges within a DVI context. So we set up a surface decomposition experiment for 14 day period. And this is just in line with DVI time frames. And at five different time points, we collected a distal phalange, which was basically in the form of a whole digit. And that ultimately provided us with samples of tissue, nail and bone. So we can see by day 14 that the body's in an advanced state of decomposition and mummified and mummification had actually occurred by about day 10 within the experiment. The experiment itself was carried out in the Australian summer. And we did actually have a couple of of 42 degree days within this 14 day time period. So 42 degrees Celsius, about 108 for our American friends. So here's just a close up of the hands at day 0.
And here's just at day 14. So, again, we can see quite extensive decomposition. So we compare the different sample types yielded. So nail, tissue, bone and the whole digit. And this also included the nail clipping versus the whole nail or the nail bed. So removing that nail bed from mummified remains was quite difficult. So while collecting that nail clipping was quite simple and amenable to an infield collection. So we found that all sample types yielded DNA profile, although tissue was unreliable. But that is in line with what was reported in the literature. We found that all samples... sorry, we suggest that the foot may be targeted to reduce the risk of extraneous DNA encountered in fingernails, for example. It was actually here that we also tried putting a little toe sample directly into a lysis buffer. The benefit of that removing all of the sample preparation steps. And it was using this rapid protocol that we were able to recover a full DNA profile.
So then we went back to our original preservative solution and applied it to toe samples, specifically we targeted two middle toe samples, which had undergone about six days of decomposition within that 14 day study. So I left the samples in solution at room temperature for eight days and at different time points I would dip a swab in and then submit that swab for routine DNA testing. So that the preservation properties of the preservative solution are well characterised, what we really wanted to test was the ability of the solution to leach that DNA. So it’s leaching properties. And the thinking was there is if we need to actually wait several days for sufficient DNA to leach, it's really not a rapid protocol.
So at every time point, for both samples we were able to recover full profiles and I've just actually recently tested the samples to test the possibility of processing them through a routine workflow in the event that the testing within the solution was unsuccessful. So at the end of those eight days, you still have basically a preserved sample that can still undergo routine testing.
Now in February 2020, I was lucky enough to be invited to carry out testing on two rapid DNA platforms at a national DVI exercise, carried out at the Training Facility for Taphonomic Experimental Research. And here we sought to assess the utility of the rapid DNA platforms for rapid profiling of compromised human remains, as well as the specifically with the in-field operation, as well as the expert and non-expert operation.
So the exercise saw six bodies decomposing for two weeks in the Australian summer and the disaster was a building collapse scenario following a terrorist incident. So consequently, there are also fragmented remains that were located around and within an exploded vehicle.
So as well as rapid DNA testing, we also tried to target sort of rapid samples, so these are really samples that are readily accessible but are still actually present within highly decomposed remains, as well as being easy to collect. So we also sought sample types that would allow a minimal sample preparation. And it was these factors that ultimately led us towards nail, tissue biopsy, hair, small bones and swabs of the actual fragmented remains.
So by choosing these samples, we would negate the need for that more invasive labour-intensive sample collection. Again if we consider femur, followed by extensive preparation and processing steps. So with regards to that minimal preparation, what I mean here is both the actual collection and the preparation. So samples that can be collected by simply picking up, cutting or swabbing. And in terms of preparation prior to the actual DNA processing, this is really referring to the removal of repeated washing steps and incubations, because it's these things that really add time to what might be defined as a rapid process. And so therefore, by using these sample types, the sample preparation can be as simple as cutting the nail into smaller pieces, rinsing debris such as soil or decomposition fluid off with water and smearing biopsy tissue onto a swab.
So we recovered DNA profiles from tissue biopsy, nail, bone marrow, hair and bone shavings. Nail and tissue biopsies do show promise, including more DNA alleles, combined with a very simple collection and preparation prior to, you know, doing the actual testing on the platform.
So sample amounts, rapid preparation protocols and tweaking that interaction with the swab in the lysis buffer will assist in optimising these sample types going forward. The study provided the first assessment of both the ANDE 6C rapid DNA system and the Rapid Hit ID system in a remote in-field application to instruct the development of rapid protocols for identification of compromised human remains. So the rapid DNA process effectively streamlines the collection, preparation, profiling and analysis, as well as eliminating that need to transport samples to the laboratory. And we're able to provide results in under two hours. So both platforms also demonstrated a utility for assisting with the rapid on-site identification and re-association of human remains. So look out for a publication on this work coming out soon.
So my final phase was to carry out testing on remains from burial samples. So here we set up a shallow grave study where we buried donors for one year and two years. And we excavated the donors longitudinally, which allowed us two time points from each of the donors for that sample collection. So we carried out the exhumation at one year, and it did actually show quite drastic differences between the donors. So one was mummified with skeletonisation in some parts. Apologies. While the other one was a lot more skeletonised, and that was despite those donors being separated by about two metres. So at two years, both donors were a lot more skeletonised, although we did actually still see some tissue remaining. But prior to this collection of samples we collected samples from a donor that have been subject to surface decomp for four years. And that was really to test the feasibility and really trial our rapid protocol. So as you can see, the bones of the distal phalanges had extensive contamination of soil and moss.
And to test these samples, we applied that same limited preparation and direct incubation in lysis buffer for two hours and 15 minutes. So the preparation was limited to wiping with bleach, water and ethanol and then either submitting it whole for two hours or hit with a hammer and then for 15 minutes. So the thinking was breaking that that bone with the hammers to break open the bone matrix and really allow that lysis buffer to get in and ultimately facilitate DNA lysis. This really took the work of Harrel and Houstein and Dai... Combined it and took it forward with our sample time. So we actually saw that the crushed bone and 15min intubation gave slightly better allelic recovery and this could ultimately reduce bone processing time from two and a half days to a few hours. So the results should be coming soon. So one thing we have observed is difficulty in recovering distal phalanges and the nail sample, especially in that shallow grave scenario. So feet in shoes have made it simpler, and it also does seem to somewhat preserve the sample within. So we'll also study differences between the distal phalanges of the hands and the feet, as well as the intra individual differences. So is the first distal phalange containing more DNA simply because it's bigger than the fifth, for example. So just in conclusion, we've been able to develop protocols that continue to make DNA testing of compromised samples faster through a variety of approaches, which is of benefit to DVI. And this includes automation, direct PCR methodology targeting sample types amenable to rapid collection and preparation, a rapid preparation of these samples itself, rapid DNA testing, platform testing and a rapid DNA lysis protocol. Thank you.
Thank you very much for that, Jeremy. As mentioned, we'll leave questions until the end of the session. So let's move straight on to our second speaker and that's Sharni Collins. She's also a PhD candidate here at UTS. She completed both a bachelor's and Honours degrees here, allowing her to specialise in forensic taphonomy, such that her research is also in collaboration with AFTER. Her research is currently focusing on the investigation of postmortem lipids in complex matrices associated with human remains, which is what she's going to speak on now.
Thanks for the introduction, Georgina, and great talk Jeremy. It's really nice to see the application of your work too. All right, so as you mentioned, I'm going to be presenting to you some of my PhD project. It is just a snapshot just for the sake of time, but I am looking at post-mortem lipid degradation and complex matrices associated with human remains. And I am a PhD candidate here. It's under the supervision of Dr. Maiken Ueland and Barbara Stewart, Associate Professor. So my field of research is in Forensic Taphonomy, and it is a unique field where we look at all of the complex processes that occur to the human body as it decomposes from soft tissues to hard tissues. Unlike what you might see on TV and in movies, it can be quite difficult to do this because there are a range of different factors that do affect it, such as scavenging activity in temperature and climate and insect activity, just to name a few of them. But what we do in taphonomy can be quite useful in aiding investigations by assisting victim identification, estimating the time since death or what we refer to as a postmortem interval, identifying cause of death, suspects and people of interest, as well as locating primary and secondary crime scenes. So it can be quite useful as a holistic approach for investigations.
So you might be thinking why and how my particular project fits into that and why I've chosen to look at lipids in complex matrices such as textiles and soil. The lipid portion is quite simple and easy to explain. So the human body in life is made up of a range of different macromolecules ranging from proteins, lipids and carbohydrates. As we decompose from our soft tissues into the hard tissues, it produces liquids and gases, and these liquids and gases in the decomposition process are primarily made up of those macromolecules. Now, the reason that I've chosen to target lipids as opposed to, say, the carbohydrates or the protein production, is primarily because quite a lot of research in agriculture and archaeology have shown that lipids persist in the natural weather environment for a long period of time. So they are a good target. And in a forensic context, that's a very good foundation for my work. In terms of the complex matrices, I will focus only for textiles for the sake of time, and I wanted to introduce to you a few Australian cases which you may or may not have heard of, depending on your demographic and geographic location. But the first one is Lynette Dawson. So she is an Australian mother of two that went missing in the 1980s. You may have heard about her story through the infamous Teacher's Pet podcast that came out a fair few years ago and she actually went missing in the 1980s and her remains are still missing up until today. And the other two are mother and daughter Karlie and Khandalyce Pearce, which were unfortunately victims of homicide and Karlie's remains were found in a skeletal state, in an outdoor environment so in a bushland forest and Khandalyce was found decomposed in a suitcase on the side of the road.
Although very tragic, the three cases, they're quite different, and that's what I wanted to draw your attention to here. So we have remains that are missing remains, the skeletal and remains that are decomposed. But all three of them had one thing in common, and that was textile evidence. So that provides a good foundation again, for my research is I'm targeting readily available physical evidence to apply a new method to gain more information. So how are my matrix complex? Will the textiles themselves range in their type. So a cotton t shirt will be very different in the chemical and physical properties as opposed to, say, polyester pants. And that makes it quite complex to start with. That will then flow into the degradation patterns. So, again, the cotton t shirt will decorate a different light compared to the recipients being that one's more natural and one synthetic. The anatomical region on the human body, so we wear t shirts on our torso and pants on our limbs, that will impact the analysis further down and also the interaction between the textiles themselves and human remains and vice versa makes it all quite a complex matrix to work with. In terms of soil, well soil is an organic compound, and it's made up of different endogenous compounds from plant and animal matter. The chemical and physical properties make it very complex to work with, as well as that interaction that I mentioned before. So between the soil and the human remains and vice versa.
So that's a good place to introduce the research facility. So we've heard quite a lot about it through the introduction from Claude and then from Jeremy again. But it is the Australian Facility of Taphonomic Experimental Research. So we refer to it as AFTER, a bit more digestible name. And it is a unique outdoor woodland environment that we can study whole human decomposition. So not only is it a unique opportunity to do this in terms of the legal and ethical ability to do so, it is the only one in the southern hemisphere. So it allows us to further the decomposition research that has previously been done overseas and in an Australian environment, as well as testing the differences between human and animal models. Because there is quite a bit of disagreement in our field at the moment as to whether or not pigs are a good model for humans. And so it allows us to test that.
So the aims and objectives of my research is looking to develop and optimise a method for the extraction and detection of post-mortem lipids in textiles and soil. So the complex matrix, as I mentioned. To then identify any trends and patterns in the postmortem lipid profile to assess viability for time since death estimations. So if we can see that the lipids are degrading in a predictable way, perhaps we can then relate that to the time since death.
And then I'll be applying that in a range of different environments that we would simulate for casework. So at the moment, we've got a surface vs buried study. So we have remains on the soil surface and then remains buried. We also are conducting pig vs human study, which again will hopefully answer some of those questions and tie up some of that disagreement in the field about pigs and humans. And then another one is a bit of a small scale study about looking at textile removal. This did come from a review, a comment on a paper that we submitted. And they wanted to know whether or not the lipid profile of a scavenged textile from human remains would then mimic the lipid profile of the textiles that remain on the human as decomposition progresses. So how do we get to that point where I can apply a method and then test on these kind of samples? It is through quite a lot of method optimization and development. Luckily, one of the first steps that I'm following is kind of like a preliminary screening method, and that's already been established for use of textiles, for example, and that's using FTIR. So this is a great way to fast track our analysis as a first step because it is simple and non-destructive, which in terms of forensics, is a really good advantage. But unfortunately, it isn't a targeted analysis system and nor is it quantitative. So it has quite a few limitations there. So that's why I'm employing a complementary technique through the use of the GC Tandem MS, which I may refer to as a GC-MS/MS or GC triple quad for the presentation, and this is a great analytical tool because it is targeted and quantitative and it also is selective and sensitive. So for our analysis, it's an excellent tool.
But in order to get to that point where I can test my sample both on that screening and confirmatory method, there's quite a few development optimisation steps for the GC triple quad, and that involves, first, optimising my extraction. So being able to actually get the lipids out of the matrix, so for the textiles, for example, I tested six different methods, some of which were already published and some of which we kind of developed along the way and actually found that a one to one ratio of acetone and chloroform in solution yielded the best results. And those results are not necessarily based on the highest abundance of the target analytes, which are the lipids in this case. But it was actually the suite of analyses that we could get. So this method allowed us to target saturated, unsaturated fatty acids as well as sterols at quite a good yield, whereas some of the other methods would only target the sterols, not the fatty acids and vice versa. And that's just based on the chemical properties of the solvent. So now that we've extracted the liquids out of our material, we want to be able to then run it for analysis, but we need to, again, optimise those steps and that's through the gas chromatograph, so that's our GC optimisation step. And this allows us for separation and detection. But we need to adjust a few things in terms of temperature, temperature ramping, flow of the gas through the column and also the column properties. And once we've got that all sorted, we then move on to our detection. So the unique ability for the GC tandem MS, or GC triple quad, is that we can use the MRM mode, which is multiple reaction monitoring mode, and that allows us to target specifically our analytes. So in this case, it's our lipids and then we can fragment them based on mass and retention time into parent and daughter ions. And these parent and daughter ions are specific to the analyte. So it allows us better selectivity and sensitivity as opposed to, say, a regular GC-MS. We were able to do this on thirty three lipids using standards. And the reason that we've got the different types of lipids in our method, we've got things like ergosterol which is a fungal sterol. We've got human specific fatty acids and sterols as well. And then we've got a range of lipids in different carbon counts. And that's ranging from larger to smaller carbon count. And the reason that we've done that is so that we can hopefully track the patterns in the degradation from larger to smaller lipids over time and then hopefully capture a more holistic view of the decomposition by having those environmental lipids in there as well.
So once we've done MS optimisation, we need to calibrate the instrument, which will then help us downstream for quantitation steps. And that's through calibrating with limit of detection and limit of quantitation again, through standards. And now and we're at the point where we can identify and quantify lipid profiles by testing on real samples.
So I want to talk you through just a bit of a snapshot from one of our surface studies just in the interest of time, and we'll be looking at cotton textiles for this specific example. So this is a general set up that we have, so our experimental is with human remains and textiles, so the textiles were then exposed to the decomposition process over time, and that is just on the soil surface laying in a supine position. So the donor is facing upwards and then our control is placed at the identical time frame in the facility without any human remains or any contamination so that we can have that as a negative control. So once we take our textile samples from the field, we then bring that into the lab for the preliminary screening step and that's using that FTIR that I mentioned before. And that will give us an infrared output, something like this. So what I've got here is three different samples stacked above each other. So on the Y axis, I've got the offset absorbance values. And on the X axis, we've got the wavenumber. And I've selected these samples just because it gives a good example over time. So we've got days zero post-mortem up until day 19. And a few things that we can do at this screening method is look at the textile degradation itself. So I want to remind you that we are looking at cotton, so we'll be looking at the cellulose region here. And that's because cotton is primarily made up of cellulose. So we're looking at any conformational changes in the shape and size over time that could indicate to us that perhaps there's any textile degradation occurring. And then the other thing that we can do at this step is look at the Carbonyl region as well as the CH stretching region, because that can indicate to us whether or not there are any lipids coming through in the sample. And we can even have a look at the transformation from triglycerols to free fatty acids. And this is based on quite a lot of literature that's already out there. So we can use that as a reference. But as I'm sure you can understand, this is quite limited. It is primarily a visual inspection method. It is great as a screening tool, but it doesn't give us any more information from that. And that's when we apply the complementary method. So we then take those samples, we section them into duplicates, extract them using that solvent that I mentioned to you before, and then we need to do an extra step through derivitisation. That's primarily because our sample being lipids, are non-volatile. And so this step allows them to be volatile so that they can be separated and detected using the gas phase.
We then inject that onto the GC-MS/MS and we get a chromatography outlook that looks something like this, and these are the same samples that I just showed you in the infrared. So this is through the GC-MS. So along our y axis, we have area and the x axis retention time in minutes, and again, I stack them from day zero to that day 19 sample. And a few things just at a glance is what you can do here is have a look at this region here. So retention time is about 12 and a half minutes and over time you can see that this peak increases quite significantly. But then what is great about having this technique is we can get more data output through using that MRM mode and we can then graph the information, something like this. So along the Y axis, we've got the area based in a log 10 scale. And that's just because the variation in all the analytes didn't allow me to show on the one graph. So we've done it in log base 10 scale. And from a glance we can have a look at that twelve and a half minute peak, which was actually the stearic acid and can see that it is one of the most abundant compounds as opposed to, say, that fungal that I mentioned to you before ergosterol, which is one of the lowest.
So overall, a bit of summary in future directions, still probably about midway through the project, so I'll continue to develop and optimise the method as we go. Already there are quite a few things that we're coming into, including saturation issues, so having to employ further dilution steps and things like that. But that's all part of developing and optimising a new method. And once we've got that all developed and optimised, we'll then hopefully have created a complimentary screening and confirmatory technique which can then hopefully assist with workflow and evidence handling individual cases and the downstream effects of that in terms of public and those that have lost people to invest it to homicide investigations and things like that will be greatly impacted.
So just a few acknowledgements as well. I wanted to thank my lab group, to Analytical Forensics, UTS CFS as well. So the Centre for Forensic Science that we have. My research was supported by the Australian Government Research Training Program and the UTS President's Scholarship. And AFTER, so a special thank you to all of our donors and their loved ones for the invaluable gift of science, because without that, none of my research would be possible. And it's a very selfless gift to do. So here are some of the references that I had on this slide, and as Georgina had mentioned before, we'll do questions at the end, but I also just popped my email here as well. So if you guys have any further questions, feel free to email me.
Thank you so much Sharni. OK, so we'll go on to our last but not least speaker Amber. Amber is also a PhD candidate here at UTS but in collaboration with the Australian Centre for Wildlife Genomics, Amber is currently developing novel forensic methods aimed at detecting illegally trafficked reptiles in transit, as well as developing a fit for purpose mitochondrial DNA test to help determine the geographic origin of seized shinglebacks. We'll now hear more about one of these novel detection methods. Over to you Amber.
Thank you for that Georgina. All right, well, I'm excited to be here with you guys in developing field and wildlife forensics. So wildlife forensic science encompasses a majority of disciplines that are used to investigate human crimes. However, in this instance, your victim is now a wild animal. And because of this, the use of these analytical techniques, including genetics, toxicology and veterinary pathology, will differ in order to determine whether or not a wildlife crime has been committed. However, as the use of this evidence will be used for prosecution, the same standards for evidence collection, analysis and handling is the same as human forensics. Wildlife crime extends to a variety of different actions. The first of which can be actions leading to the morbidity or the mortality of an animal, including poisoning or illegal poaching or unlawful killing. This can also extend to the harassment of protected species, as well as animal welfare issues and the illegal international or domestic trade of wildlife, which is also known as the illegal wildlife trade. The illegal wildlife trade include the trading of both deceased and living protected specimens and can be both domestic and international. Not only is this trade cruel, but it also it has inherent biosecurity risks. Anyone that has been in quarantine this year can thank a zoonotic disease for that. As seventy five percent of emerging diseases are zoonotic, this trade only increases our risk of exposure. Additionally, the illegal wildlife trade has long term ecosystem impacts, both from the source country from where the animal was taken and removed, as well as the introduction to a new country where if the live animal was released, it could become an invasive species.
Additionally, more research is showing that the illegal wildlife trade is also associated with other criminal activity, such as the illegal drug trade and the trade in illegal arms. In order to prosecute a crime related to the illegal wildlife trade, you generally need to gain information regarding the species identity of what's being traded and under what regulations that animal is protected. It's also important to determine the heritage or the parentage of this animal. This is because from a compliance perspective, you want to ensure that the breeder has the correct regulations to breed the animal or if this animal was sourced from the wild. It's also very important to determine the geographic origin of the animal. As everyone knows, animals don't really listen to different borders and can extend across different countries and their species range. So determining the geographic origin of a seized animal can help develop poaching hotspot intelligence, and it can also help develop which jurisdiction this crime needs to be prosecuted in. Lesser related to the trade of live animals, but instead the trade of specimens such as ivory is determining the age of that specimen because there are times where you can hold an antique specimen and it's not considered illegal, as where a recently poached piece of ivory would hold a higher punishment.
Of the illegally trafficked live animals, reptiles are the most highly traded live taxa. Unfortunately, over one third of the described reptile species are found on this trade, and less than a quarter of these species have international regulations. This is particularly concerning because the value of a reptile in the exotic pet trade is its rarity. This means that species that are either recently described to science or species that are inherently hard to find, such as this beautiful, earless monitor lizard from Borneo are quite frequently trafficked. Because of this, the international trade in illegal poaching of these animals has unknown impacts not only to the animals themselves, but the ecosystems and potentially what diseases they are carrying with them. The current methods used to detect illegally trafficked wildlife include visual examination of packages, X-ray detection and the use of wildlife detection dogs that are able to detect trafficked animals based off of their odour. However, traffickers, especially if they're good, can prevent detection through binding the animal and preventing its movement and vocalisations. By placing animals in different containers, which can complicate X-ray detection or through using more potent adulterating odours to confuse wildlife detection dogs. However, of the available methods, there is huge benefit and odorant detection. Odours are comprised of volatile organic compounds, which I'll refer to as VOCs for the duration of this talk and from a living animal, they're being constantly produced and released. This has both ecological and biological benefits for the animal as they will use it for identification purposes, territory marketing and environmental exploration. VOCs are also generally small molecules, so they are able to penetrate both porous and semi porous packagings, of which the reptiles are generally shipped in. Also, the profiling and detection of VOCs is nondestructive and noninvasive, so it has the opportunity to be implemented into current workflows.
In order to profile VOCs in a forensic context, we look at the collective VOC profile known as a volatilome, to see what odours or VOCs the target is producing. We will then identify either individual or suites of compounds that are unique to the target and can be used for identification or detection purposes. This has been used in many fields of forensic science, including the detection and location of human remains. There's also been research in using verses produced by decomposing human cadavers to help establish a timeline since death. VOCs have also been used to detect and identify narcotics, explosives and ignitable liquid residues that are associated with arson. More recently, there has been an application of VOCs in wildlife forensics where the volatiles produced by trafficked items such as ivory were able to be successfully distinguished from illegally trafficked bait, which include cow bone. From a live animal perspective, we expect that the volatilome will be influenced by stable primary factors, unstable secondary biotic factors and tertiary abiotic factors. Now, I can't tell you how many reptiles are wearing perfume. So instead we include this as any external application that is unrelated to the biology of that organism.
From a wildlife forensic perspective, we're more interested in targeting compounds that are related to genetics, diet and microbiome, because we suspect that by targeting these compounds, we'll be able to answer questions that are more related to wildlife forensic investigations. Because of this, the aim of my project, at least the first chapter, is to optimise reptile volatilome collection and analysis methods, again targeting different compounds related to genetics and diet. And the second aim was to determine if we could extract any more granular information from the volatilome, such as species specific biomarkers that can be used for species identification or biomarkers that are shared across reptile species that can be used for detection purposes. We were also interested in seeing if we could tell the difference between an animal that has been captive bred or caught from the wild.
The first step of optimisation is determining which sorbent is best to collect the odours that are being produced by a reptile. To do this, we use two sorbents. The first is a general sorbent called Tenax TA. Next to that is a considered a weak sorbent, and the second sorbent combines Tenax TA with an additional stronger sorbent to extract to extend the carbon range. In order to do this, we acclimated one male shingleback for 20 minutes and did three sampling intervals in triplicate at 10, 15 and 20 minute intervals, I should say, for the rest of the study between each interval and between each animal, we washed the container with acetone and vented with a hair dryer. We did this to remove any VOCs from a previous animal in the container, as well as to determine what volatiles are being produced by the container and not the animal itself.
Our results from here showed that the first sorbent or pure Tenax TA always retrieved more compounds than the second sorbent. However, from a forensic perspective, more compounds in this case was not more informative. Instead, sorbent one was too sensitive to the changing biological conditions of the animal, as well as changing environmental conditions. Sorbent 2 with the addition of that stronger sorbent was less influenced by these factors and then was thus selected by the as the forensically relevant sorbent for the rest of this study. The second optimisation parameter was the acclimation time or how long the animal was acclimated in the container. Not only was this to produce VOCs, but from my picture here, each shape represents a different VOC that has a different molecular weight and volatility. So this acclimation time also allowed for these compounds to equilibrate within the sampling container. In order to do this, we sampled one male and one female shingleback with 15, 20 and 25 minute acclimation periods. Through this, we found that the 20 minute sampling, ah acclimation interval was the best at creating a stable profile. The next parameter was the sampling interval or the time spent collecting a reproducible profile over the course of three replicates.
In order to do this, we used one male and one female shingleback and sampled in triplicate every 10 and 15 minute sampling intervals, you will notice that we removed the 20 minute sampling interval that we used during our sorbent optimisation. And this is because the shinglebacks expressed escape behaviour and we were not able to collect triplicates using the 20 minute sampling interval. Through this, we found that the most reproducible profile was collected at the 15 minute sampling interval. All of our analysis was conducted using a thermal desorption and comprehensive gas chromatography. So the first step we had to do here was determine how can we retrieve all the compounds that we had just sample from the animal. We optimised the thermal desorption parameters and found that a moderate flow, a low, cold, trapped temperature and a moderate cold trapped absorption flow was the best at retrieving compounds collected from the shingleback. We also want to extend our collection analysis parameters to different species of reptiles, and we suspected that the volatilomes of each reptile would differ for species. Because of this, we wanted to optimise the selection of columns that are used in comprehensive gas chromatography in order to accommodate these additional species. In traditional gas chromatography, you typically only have one dimension in which to separate compounds based off of the chemical property.
In this case, our first dimension is based off of the volatility of that compound. However, the added benefit of comprehensive gas chromatography is the second dimension where you're able to separate compounds on an additional property in which this case's polarity based allows us to see a more robust profile and determine even more compounds that are being produced by the animal. Here are my two best chromatograms from the shingleback lizard using each column set. On the x axis, is the separation time in that first dimension and on the Y axis is the separation into the second dimension. Each dot represents a detected compound where the black represents a low relative abundance and the red it represents a high relative abundance. You can see that both column sets here. We're able to separate compounds in the both the first and second dimension. However, column set one appears to have more compounds with different relative abundances. We wanted to see how the difference in abundance and our identification of compounds affected the total profile of each animal that we analysed. Using column set one you can see that there is a clear difference in volatilome profiles between the shingleback lizard, Eastern Bluetongue Lizard and the children's python. However, using column set three where we have reduced sensitivity, you can see that these profiles start to look very similar and we have an increase of unknown or unidentified compounds. Because of this, we selected column set one as our optimal column set for the rest of our analysis.
We additionally ran a principal component analysis to get a vague idea of how our collection analysis methods were working in a forensic context and using column set one we could see that there was a close clustering of each replicant that we took from each species. And we're starting to see that there is species differentiation between the volatilome each animal that we sampled. However, when we lost the sensitivity using column set three, we can see that there is more separation between our replicates in each animal and starting to see some species overlap. Again, this highlighted that column set one was best for the trace analysis of reptile volatilomes. However, it should be known due to the high dimensionality of our data and the logarithmic changes in our abundances, we are developing a more rigorous statistical method in order to determine the influence of these compounds, as well as how important they are to the species identification or other information related to reptiles.
In conclusion, through this study, we optimise a volatilome collection and analysis method for reptiles, which will set the foundation for future forensic research. Preliminarily, we are seeing that there are dietary influences to each profile, which also sets the stage for future forensic investigations, not only relating species specificity, but again, perhaps geographic origin of seized animals. We have only just begun, and so we are collecting a much larger data set in order to answer some more of our questions, in order to determine if we can tell if a species is captive bred or wild caught, we are sampling both captive and wild shingleback lizards as a model.
We're extending this to sample shinglebacks across their species range. This will allow us to see how a profile changes over space and time, as well as help determine if the profiles differ based off of geographic location, diet or species. And again, we're sampling approximately 30 different reptile species to see how specific these profiles are or and to what taxonomic differentiation we can distinguish animals from each other. We're also doing this to determine reptile specific biomarkers that can be used to train an electronic nose to detect illegally trafficked wildlife and for reptiles in transit. This work could not have been completed without the support of our funders, including the RSPCA, SWFS, which is the Society of Wildlife Forensic Science, the Holdsworth Research Endowment. I'm not sure if you could tell with my accent, but I am international. So the UTS International Scholarship and President Scholarship, as well as the support of the Australian Museum Research Institute, I'm very thankful for all of my lab members, both within UTS and the Australian Museum, as well as all of my supervisors who have gone in the field with me and have been trapped in polar vortexes or floods or fires. We've also had a lot of support, both from Featherdale Reptile Park and the Australian Reptile Park.
I realize we were saving questions for the end, but this is a another way to show you guys a cute picture of a shingleback, and I guess I'll leave it to Georgina for any questions.
Thank you very much Amber.
Drugs and Drugs Intelligence parallel session
Crossing Forensic Borders Drugs and Drugs Intelligence Parallel Session, hosted by Associate Professor Shanlin Fu and Dr Marie Morelato and featuring speakers Madysen Elbourne, Eleanor Finch and Ana Popovic.

Crossing Forensic Borders
Drugs and Drugs Intelligence Parallel Session
Morning. Good morning, good afternoon, good evening, depending on where you are. I would like to welcome you all to attend the Drugs and Drug Intelligence section. I'm Dr Shanlin Fu, Professor in Forensic Toxicology at the Centre for Forensic Science UTS. I'll be chairing this session and I'll be assisted by co-chair Dr Marie Morelato. Thank you, Marie. Just a couple of housekeeping messages before the presentation. I ask everyone to keep your microphone on mute during the presentation. I also please ask you to hold your question at the end of all three presentations. You may type your question to the chat. And I would try to get them to answer them later. Or you send a message to the chat indicating that you want to ask a question at the question time and I will call you and you can ask your question using your microphone. Again, and this session is recorded, so if you have any concerns, you can contact the UTS department and you can send and express it there. OK, let's start our presentation today. The first speaker is Madysen Elbourne. Madysen completed her undergraduate study of forensic science and chemistry in 2019 at UTS. And she completed her Honours project last year working on an anti-doping project in collaboration with Racing New South Wales. So Madysen's starting a PhD this year, furthering her Honours research. Today, Madysen will present part of her Honours research findings. So the title of her talk is GC-MS Profiling of Equine Urine for Longitudinal Assessments. Now the floor is yours, Madysen. Thank you.
Good morning. My name is Madysen as Shalin said. And today I'll be presenting some of my Honours project findings, which are on the topic of the GC-MS profiling of equine urine for longitudinal assessments. My supervisors for this project were Shanlin Fu, Adam Cawley and Chris Bowen. And this project was in collaboration with Racing New South Wales and Shimadzu. So Racing New South Wales has established the world's first thoroughbred equine biological passport, or EVP, for short, and this is to perform longitudinal profiling of equine urine samples collected from specific horses over time. The essential analysis of biomarkers by targeted analysis and untargeted feature extraction is applied to discern pharmaceutical manipulation from physiological variations. It has been discovered that dopamine in this section here, may be a target for illegal manipulation, and an EVP will allow the analysis of the individual horse biomarkers and detect any misuse of these doping agents. So what's a biomarker? Well, a biomarker is defined by Teale as 'any measurable parameter altered as a result of a challenge to the individual's system'. When endogenous compounds are abused or altered, it can be difficult to detect as their presence in the urine sample isn't unusual. This is where the use of biomarkers is especially important. Biomarkers can be analysed to more accurately detect unnatural levels of substances that may have been doped, performance enhancement or suppression. Endogenous reference compounds or ERCs can also be utilised to detect these abnormal levels. So my project mainly focuses on the use of levodopa and dopamine since the discovery of the benefits of levodopa in human Parkinson's disease patients, it's been suspected to have been misused by the sporting population. In humans, levodopa works by increasing the levels of dopamine in the system, which causes increased freedom of movement in the limbs and the trunk of the body. Such improvements in the locomotive activity is assumed to also be the motivation for the illegal use of the substance in racehorses.
So 3-methoxytyramine, or 3MT, was the main compound that I analysed and tyramine just here was also analysed and assessed as a possible ERC to create a ratio with 3MT. The benefit of using an ERC is to improve the distinction between body and bottle. And this also accounts for the physiological variations. Previous research into the correlation between dopamine and 3MT has resulted in a urinary threshold of four thousand nanograms per mL put in place. This threshold, however, is pretty conservative and it doesn't account for people manipulating within the levels to stay within that threshold.
So my project aims were firstly to create a method developed for the routine analysis of samples for the EBP. Secondly, it was a review of the 3MT levels in equine urine from New South Wales horses in a reference population. And then lastly, the investigation of ERCs to be used for intelligence purposes for longitudinal profiling. And in this case, it was tyramine.
So my sample preparation, three mLs of urine was pH adjusted to between four point nine to five point five and then an enzyme solution d4-3-MT, as my internal standard were added. A buffer of phosphate was also added. And then the sample was incubated at 37 degrees Celsius for 12 hours. The next morning it was then centrifuged at three thousand RPM for ten minutes. Solid phase extraction is also then commenced using the UCT Xtrackt cartridges which are conditioned, washed, and then the basic fraction is eluted using 3% ammonia, 0.5% methanol in an ethyl aetate solution. 0.5mLs of that is then aloquotted and derivatised using 15 microlitres of methanoic HCl, 25 microlitres of acetic anhydride and 25 microlitres of pyridine. That sample is then finally analysed using the Shimadzu QP2020 and NX GC-MS.
So, GC-MS analysis of .. sorry, all these samples, were analysed using the full scan mode and electron impact ionisation with the parameters you see on the screen and also statistical analysis was done using Microsoft Excel and MATLAB from MathWorks. So method validation was also completed in the following areas, sensitivity, selectivity and specificity, linearity, accuracy and precision, recovery, matrix effects, dilution and stability. And all areas were considered sound in relation to the parameters that are usually employed by forensic toxicologists. An external quality control sample was also used, and that's also called an EQC, sorry. And these samples tested the fit for purpose of the method that I implemented. 12 batches were used over the course of four months, which allowed for the verification of the method using this plot here. And these peaks just give you an idea of the type of peaks that I was looking for and at what concentrations, for 3-MT and tyramine over the course of my project.
So over the total course of my project, a total of two thousand four hundred and ninety six samples were analysed and the data was also collected per the sample type, whether it was pre-race or post-race, the breed, whether it was a thoroughbred or standardbred horse, the gender, whether it was gelding, female or male. And then finally, the pH level of the urine sample. So looking at reference population data, for 3-MT in urine, the distribution appeared relatively uniform for an approximate range of about 50 nanograms per mL to 400 nanograms. This resulted in a mean of 231 nanograms per mL and a median of 217 nanograms. These values were not considered to be different, with a measure of approximately 6.5 percent. This data set also gave us a near outlier just here of 539 nanograms per mL and a far outlier of 776 nanograms per mL. So then looking at tyramine in the urine, these current values very obviously skewed and it's reflected in a mean of 663 nanograms per mL and a median of 526 nanograms per mL, and these values had a relative difference of approximately 26 percent. So the use of tyramine as an ERC was what we desired.
We needed to look at the ratio between the two of them. So then looking at that data, we can see from the distribution values that it is also skewed, which is reflected in a mean of 0.55 And a meeting of 0.44. And this had a relative difference of 25 percent. If we then log transform the data, the distribution actually exhibits bimodal quality just down here, which is due to samples with high tyramine levels. So there is potential for a parametric distribution to exist, with a log 3-MT/tyramine values which are greater than negative 0.8. That equates to an untransformed ratio of 0.16. And the inference is supported by this normal distribution, sorry, normal probability plot just here and highlighted by the red box.
So when we review the frequency distribution of the log transformed 3-MT to tyramine values that are between 0.16 and 3.98 and the distribution plot on the left, it provides a good approximation of a parametric behaviour for the ratio values for a one tailed approach to higher values. The reason for being able to neglect the data which is below -0.8, or 0.16 untransformed, is because we're interested in the higher ratios rather than the lower ones. And these ratios weren't really relevant for this project. This selected log transformed distribution overall represented eighty six percent of my data, and it enables the use of parametric statistics to estimate a reference limit.
So my population limits, we can propose an upper outlier limit of 539 nanograms per mL from the non parametric 3-MT reference data and then also another intelligence limit of 776 nanograms per mL. And then from the ratio data that I just showed you, we can propose an upper ratio limit of 5.26, and this is with a 99.99% confidence interval. So over the course of this project, two administration studies were conducted and both studies used Sinemet as their type. The only difference between these two studies is that Horse 1 was conducted by Singapore Turf Club and Horse 2 was conducted by Racing New South Wales. The other difference is that Horse 1 only had five samples collected, whereas Horse 2 had a total of 19. So we start with Horse 1. Looking at the data, there are two main limitations for this experiment, as I mentioned earlier, only five samples we collected. So there are no samples collected pre admin at time zero and then none post twenty four hours. The other thing I should mention is the red line right at the top, here at the 4000 mark is our current 3-MT threshold of 4000 nanograms per mL. Therefore, obviously the peak administration of this horse does not reach this threshold and therefore this horse would not have been detected with the current methods for doping analysis. From this data, we can also see that the upper intelligence limit of 776 nanograms per mL, which is the orange dotted line, could also provide an estimated post administration period of approximately twenty-two hours, just where it cuts here. And this is an estimate though, because of the aforementioned limitations earlier. And if we zoom in to the top corner to look at time in between the four and eight hour points, we can also see that there is a suppression trend occurring which is useful for our ERC.
So looking at the ratio between the two of them, we can see that thirty two is the ratio is the peak at the six hour point and the estimated post administration period of approximately twenty two hours is also consistent with 3-MT on the previous slide. So it cuts roughly of our ratio here. And then so looking at horse number two, the same limitation applies of no samples taken between eight and twenty four hours for this administration as well. However, there are a lot more samples, pre administration and then prior the twenty four hour points. It's also quite obvious to see that this horse does reach the 4000 nanograms mL threshold at its peak max at four hours. This is, however, an estimated value. But if we zoom in to the intelligence limit again of 776 nanograms per mL, we can also see that it cuts at approximately twenty two hours again. And then looking at timing, it also shows a surprising trend similar to Horse 1.
Now, looking at the ratio between the two. The only difference between this one and Horse 1 is that we have a much higher peak ratio of 290.5 compared to 32 in Horse 1's data. The ratio for this one does cut for the post administration period at around between twenty two to twenty four, it's kind of hard to tell in this one because of that lack of data between the eight and twenty four hours.
So looking at our McIntosh model, the EBP profile utilises a formula which is derived by McIntosh in his two papers on cancer research, the formula uses either the sum or the difference, allowing for the individual's upper or lower reference limits, or IRLs, to be derived. The IRLs are derived using the parametric empirical Bayes method developed for monitoring biomarkers in cancer diagnosis. This model is also used as the basis of the human antidoping detection. The way this ratio line works is that as more values of the horse are put into the profile, the weight of the individual variance goes up and the population variance goes down, which is super important. So looking at a theoretical model for this, this model mimics consistent twenty four hour sampling over the course of nine days using pre administration data samples from Horse 2 that you saw earlier. And the results show that after twenty four hours of administration at this endpoint here, 3.7, the doping of Sinemet is still detectable using this method and the McIntosh formula, which is unlike the proposed reference population ratio limit that I mentioned earlier, that we were able to adapt, which was only able to detect up to the 22 hour line.
So this IRL formula, the red dotted line, takes approximately three data points, as you can see here, from the individual to create a fairly accurate baseline limit, which is supported more by the individual's data other than the general population data.
So finally, to conclude, I was able to propose an upper outlier limit of 539 nanograms per mL and an upper intelligence limit of 776 nanograms per mL, and also a proposed ratio limit of 5.26. There was also potential negative feedback or suppression that we noticed from tyramine when levodopa was administered. And this supports the suitability of timing as a possible ERC for an intelligence based antidoping strategy. The further works for my project, is still the federal collection of reference population and a comparison of tyramine in spring to autumn, which could look at some seasonal variations that I noticed in my data. Investigation of tyrosine loading, which can account for some of those high tyramine levels, a continued collection of EVP samples, and then finally exploring a chemometric for feature extraction. I'd like to acknowledge the UTS Drug and Toxicology Research Group and all of the staff at the Australian Racing Forensic Laboratory. Thank you very much.
Thank you Madysen. If you could please stop sharing your screen and I'll call up the next speaker, Eleanor Finch. Eleanor is also a graduate of the UTS Forensic Science Program 2020 Honours program and specialised in forensic toxicology and drug intelligence. And she currently works at the New South Wales Forensic Analytical Science Service, a leading forensic science organisation of the New South Wales state government. Today, Eleanor will present part of her own research findings on improving our knowledge of drug usage through the analysis of used injecting paraphernalia. Thank you. Eleanor.
So yeah, my project that I was doing last year was improving our knowledge of drug usage by analysing used injecting paraphernalia. And that was done in association with UTS, as well as the Uniting Medically Supervised Injecting Centre over in Kings Cross in Sydney.
So the global drug problem is a multifaceted problem that surrounds the number of drug users globally of both illicit and prescription drugs, the dangers associated with risky use and the morbidity and mortality associated with use. Therefore, the drug policy that's currently in place in Australia comprises of three different pillars aiming to lower these problems. Now, I'm not familiar with the American and European situation on drug policy, so I am just going to focus on the Australian one for now. But I do hope that the research I'm presenting will be useful for America and Europe. So the main pillars were harm reduction, demand reduction and supply reduction. And for this type of project, harm reduction is important because it doesn't aim to completely lower usage, but aiming to reduce any of the harms associated with drug usage. And in order to do so, we need to monitor the usage of people who inject drugs and evaluate harm reduction strategies that can be put in place. And some of these monitoring methods include self reporting by people who inject drugs, wastewater analysis, chemical profiling of police seizures, and a new style of project which is used injecting paraphernalia analysis. So there have been previous studies conducted, a majority of this research has been done over in Europe, both in single countries such as Switzerland, France and Hungary, as well as a European wide study that went across six different countries.
So these projects, they were developing the methods that can be put in place, determining how viable it was for both single country studies as well as global and European wide intelligence, as well as understanding the initial trends that they can use to compare back to. And they discovered that you can monitor it in a pretty tiny manner and, pretty fast way. You can compare geographical locations, discover any new drug patterns or drug trends that emerge. And it's a complementary datasource to existing monitoring methods. Now, there's only been one study done in Australia, and that was by Elladele Lefrancois back in 2019. She did realise that it was a new and underdeveloped data source in Australia that needed a lot more research to become similar to the European understanding of the drug market. But that study proved that it was a feasible project style for Australia, it just needed to be up scaled.
So the aims of this project from last year then were gathering a longitudinal and objective picture of all the substances injected at MSIC in Sydney, Australia. And the objectives were using, gas chromatography mass spectrometry to determine all substances injected by clients, comparing these results back to the self reports, see whether or not the clients were aware of what drugs they were injecting, comparing the 2020 results back to 2019 to see any changes between those two years and to detect any new substances or new practices that might be harmful for people who inject drugs.
So the method that I went through, so we collected seven days worth of bins over the entire facility, collecting two hundred and ten syringes, needed to triage out those syringes from any of the other material that's shown in that picture, including the bands, spoons, any of the bottles and all of the biological waste. Sample preparation is very easy. We just pumped a mL of methanol through the syringes and then added an internal standard and put it into the GC. It's a very simple method that has been established over the last couple of years. And then we identify any of the main psychoactive substances. So these are the main drug that showed up in this syringe and it didn't mean that it was the main drugs themselves, it could have been some metabolites, but they were the only substances that were found in high enough percentages of that syringe and any syringes that came back with no drugs, they were retested to ensure that that was actually valid results. And Microsoft Excel was used to manipulate the data, but there was no quantitative analysis possible due to the fact that there was possibly blood in the syringes that could show up old results or because all two hundred and ten syringes were... They weren't single syringes, so syringes could be contaminated with other ones. And then we compared that to the self reports in 2019 data.
So the typical sort of chromatograms you were getting for methamphetamine was showing up at around six minutes and heroin usually showed up as a tri-peak of diacetylmorphine and 6-monoacetomorphine, and then 6-acetylcodeine. So they were pretty standard, luckily. So the positive syringes, so fifty three syringes came up with no results. So those ones were eliminated from the study as it's possible that they were either... That users had used multiple syringes in a single injection because a syringe broke or they needed to prepare some samples. So we discounted any of the negative syringes. So these are the results from the positive syringes. So as you can see, heroin and methamphetamine, the more most commonly identified drugs, followed then by mixed drugs. And usually these mixes were two or... One of them had a three mix, and oxycodone. So I was saying the mixes, the most common mix that was found with methamphetamine and heroin, and then there were a couple of mixes with Benadryl and methoxyphenamine. We did have two new psychoactive substances show up in these mixes and it was interesting to see them there. However, it's a little bit hard to say whether or not they were mixed intentionally by the clients when they were visiting MSIC or if they showed up in the blood residues from previous injections or previous other forms of administration. But it was still interesting and it was good to inform MSIC that people are using multiple drugs in a close enough time period so that they'll show up in this analysis.
So the adulterants that were found, pretty standard, that was caffeine and paracetamol, DNPA and ergotamine was found in a fentanyl syringe. And again, they could have been part of the blood, the DNPA and ergotamine, but they came up in a high enough percentage of that syringe that they did need to be identified here.
So comparing back to the self reports of what the clients said they were going to inject. So it was pretty similar to what they were going to inject, the top four groups that we identified in the analysis were the same top four that were self identified. And the only issues was just that the heroin self identification was a lot higher than what was identified in GC-MS as you can see that there was 6-acetylcodeine identified quite a lot in the 2020 study where there was no diacetylmorphine, there was only 6-acetylcodeine and there was some morphine and codeine that was found in the study that wasn't self-identified. This shows that either people are identifying that they were going to take heroin, but the drugs were actually these other ones, or that the because of the COVID-19 impact in the Australian border shutting about a week before this study took place, it's possible that heroin coming into the country had either stopped or started becoming more impure, or dealers in the country were starting to substitute drugs for other ones. And so the heroin was starting to get less and less pure. So these other drugs are starting to show up instead.
So the mixes were a lot higher in the 2020 results rather than the self identification. Again, either polydrug usage was a lot higher and people are identifying, or the 2019 COVID impact that there was some premix drugs that were starting to come into market. And there was a fentanyl syringe detected in 2020. It's important because it does show that this method can detect fentanyl, but because there was one syringe found with fentanyl on the same day that someone identified fentanyl, it shows that it was just able to identify a substance that was there, rather than identifying any fentanyl adulteration in heroin or other drug samples, which is very good news for Australia. And there was two drugs, alprazolam and milnacipran, were found in single syringes. It was possible that these were identified as another drug, but it's also possible they were just present in blood residues as these aren't known drugs of abuse in Australia. So comparing back to 2019, so again, heroin and methamphetamine were the top two drugs, both in 2019 and 2020. And based on the self reporting data from three hundred sixty five days before the 2019 study, it is known that heroin and methamphetamine are the two top drugs of choice for injections at MSIC. And then apart from hydromorphone, which wasn't detected, or wasn't self reported back in 2019, all the drugs that were detected in 2019 but also detected 2020, which shows that trends are pretty similar between those two studies. And again, the mixes shown in 2020 were at a lot higher percentage than 2019, it is possible that over the course of the year polydrug usage had increased or that the premix drugs had also increased between those two times due to COVID. Or it's also because we were needing to leave the syringes for quite a while after we collected them to just make sure that any COVID-19 virus had been eliminated. And it's possible that has maked it a bit more contaminated than we would have liked. And so, again, like the self report to the 2020, there was no syringes in the 2019 study that showed up only 6-acetylcodeine without heroin or morphine or codeine. So it shows that it's likely that the heroin purity of 2020 was lower than that of 2019, or that heroin is starting to get a bit more substituted because of the COVID-19 impact. And fentanyl, it was detected in 2020, but you can see it wasn't detected in 2019, luckily that doesn't show anything majorly drastically scary for drug users at MSIC, it just meant that there was no one in 2019 that self-reported fentanyl so we weren't expecting to see it. And the only one that we said showed up in 2020 was the same day someone self reporting. That has changed, however apparently as of November-ish that we have started to see fentanyl-laced heroin starting to show up in the Australian market. So future works definitely need to look into that.
So the conclusions. So this study did show that the pilot study was repeatable and we were able to determine any drug similarities and differences over a one year period. The self report data, it mostly aligned with what substances were actually detected. And heroin and methamphetamine, those two drugs of choice were unchanged from 2019 to 2020. But we have kind of shown that the heroin has possibly lower purity in 2020 than it did in 2019 in Australia. And there were some new and dangerous drug patterns detected. Whether or not clients were aware of this would then be a further study. And these sorts of dangerous patterns included some new psychoactive substances starting to show up, as well as an increase in drug mixes. But the project is justifiable for continuation and increasing the implementation of it. So we're doing some more future works. So the most important one of this is doing a COVID-19 study. So because of Australian not producing its own heroin and the majority of drugs needing to be imported, the border closures that started up on the 23rd of March is going to start having a major impact on drug supply and usage within the country. There are some bins that have been collected from MSIC in November as well as July. And so further works need to start analysing those to see the impacts of the main drugs as well as the adulterants over the last couple of months and future months to see exactly what changes. It would be helpful to increase any of the data collected from clients to see whether or not they are sticking to opioid treatment programs, etc., as well as increasing any collection sources beyond just the Sydney supervised injecting facility, as there is one down in Melbourne that only recently opened. So it would be good to move this project as well down to Melbourne, and moving into the needle and syringe exchange programs which are countrywide. And possibly also then the overdose syringe testing. So instead of analysing the syringes of everyone, also focusing just on those who overdose to see if it can be attributed to the drugs or adulterants' presence.
So I'd like to give a biggest thank you to all of the UTS staff, especially those of the technical staff who helped out a lot. All the wonderful staff at MSIC who became the taxi service for all the bins and all anonymous participants in the study as no one, no clients visiting MSIC denied participation in the study. Thank you.
Thank you Eleanor. And so the next presenter is Ana Popovich. Ana is a recent PhD graduate who completed her study last year. Unfortunately, due to work commitments, Ana cannot be here to present at this particular time. However, Ana recorded her presentation and I will ask our co-chair, Marie, to play the video for you. Thank you Marie.
Thanks Shanlin.
All right, well, my name is Ana Popovic, and I'll be giving you talk about the research I conducted during my PhD. I undertook a project in the field of forensic intelligence with a particular focus on understanding methylamphetamine markets in an Australian context.
In recent years, the spectrum of substances available on the drug market has widened considerably, with the persistence of traditional drugs and the emergence of new psychoactive substances every year. These two reports cover the evolution of the drug problem in great detail. The amphetamine type stimulant market in particular, poses a significant threat to Australia in the form of organised crime. Amphetamine type stimulants, or ATS, are a group of central nervous system stimulants, which include amphetamine, methamphetamine and MDMA. Global ATS seizures in 2018 reached 265 tons, which is the highest amount on record. The number of national ATS seizures remains high and relatively stable, with 11.2 tons of ATS being seized in 2017 to 18. So how do we combat this growing problem? Traditionally, the predominant role of forensic science was to generate evidence for court. It's often the case that forensic case data is used reactively and not to its full potential. Using a reactive approach, useful information is hidden in massive data which limits its ability to link specimens or understand drug models. A modern approach to combating the drug problem is using forensic intelligence. Forensic intelligence can be defined as the accurate, timely and useful processing of forensic case data. The forensic intelligence process can be applied to any type of trace. In this instance, it's drug profiles. Drug profiles can be compared against each other and their result evaluated to provide useful information that can be used for informing decisions.
The basis of the work conducted during my PhD stem from previous work. This framework here outlines the entirety of the forensic intelligence process. And as different aspects are conducted by different individuals or organisations, I've outlined in red, the area of the forensic intelligence process that my research focused on. The constant evolution of illicit drug markets, it's necessary to gain as much knowledge about them to disrupt or reduce their impact. There's been a huge effort to combat this problem through forensic intelligence and illicit drug profiling. In Australia, there are two main programs which are dedicated to illicit drug profiling, the Australian Illicit Drug Intelligence Program, or ADIP, focuses on border detections. While the Enhanced National Intelligence Picture on Illicit Drugs, or ENIPID, was set up to provide a snapshot of the drug situation across states and territories. For example, drug samples collected in New South Wales are analysed at the Forensic Analytical Science Service. Now FASS doesn't provide profiling. Their workflow only encompasses creating case reports for New South Wales police. However, the ENIPID program provides resources to allow state level seizures to be analysed and profiled, according to the National Measurement Institute workflow, with the target of generating intelligence products for the Australian Federal Police.
Intelligence products can be generated to aid decision making at various levels. Additionally, the concept of forensic intelligence in an intelligence-led policing context is to go beyond the traditional roles of forensic science and its heavy commitment to the court. There's a desire to focus on generating intelligence for proactive policing, supporting an intelligence-led model, rather than focusing on individual offenders. These different levels have varying outcomes and in particular for drug intelligence, these outcomes could be to identify links between individual offenders, drug trends or system vulnerability.
Recently, there's been an emergence of intelligence work conducted at UTS. This list of 10 citations represents some of the efforts by UTS researchers since 2010. Following on from some of the work conducted in these articles, my PhD research focused on understanding drug markets in an Australian context. Traditionally, drug profiling involves numerous analytical techniques, which can be costly and time consuming. The first objective of my work looked at the possibility to prioritise analytical techniques to achieve accurate and useful results in a timely manner. The second objective aimed to gain an understanding of illicit drug markets at a strategic and potentially operational level through various analyses. Finally, to enhance the way all this information is communicated, the last objective looked at developing a visualisation tool that would help improve the operational workflow of the Australian Federal Police.
The methamphetamine state samples acquired from the Australian Federal Police were part of the ENPID project. As a reminder, this project takes illicit drugs seized by state police and subjects them to the same chemical analysis as border seizures, with the aim of better representing the national illicit drug market. The current dataset features all Australian states and territories except Queensland during 2011 to 2016. It should be noted that each state or territory chooses which seizures are sent through to the ENIPID project. Therefore, the issue of representativeness will affect some of the analysis in this research. Additionally, each state and territory started contributing to the ENIPID project at different times. Amongst the case data relating to each specimen, for example, they analysed. And location of seizure. There is also data relating to three different methamphetamine profiles. These include GC-MS, IRMS and chiral profiles. As these profiles reveal information about a specimen in different parts of its production, we cannot simply merge these three profiles together. Additionally, these analytical techniques are not timely and can be costly. Therefore, the first objective looked at prioritising one of these profiles for subsequent analysis. To achieve this, it was necessary to determine how indiscriminative each methamphetamine profile type was.
The most discriminative profile type will be the one with the greater separation between the intra-variability and the inter-variability, or the linked and unlinked sample populations. Now two specimens are considered to be linked if they come from the same seizure, while specimens are considered to be unlinked if they come from different seizures. When you have a new specimen, you want to be... You want your model to be able to tell you accurately whether or not it is linked to another specimen. To evaluate this linkage properly, we first need to train our model.
In the initial stages of training our model we need to choose appropriate target variables in our data. When analysing illicit drug specimens, it's often found there are variables present other than the illicit compound. These variables take the form of impurities, adulterants, wetting agents, precursors and/or solvents, all in different ratios. It's important to note that the selection of target variables will highly influence the results obtained from the chemometric models subsequently applied. There is a criteria that needs to be satisfied when choosing target variables, which is on the screen. After target variables were chosen, numerical data was extracted from each chromatogram to form a matrix. Subsequently, three treatments are employed as a measure to reduce instrumental influence and therefore an important part of the overall profiling process. The most common pre-treatments are outlined in this table. For a more thorough list, please refer to the reference. Previous research by Morelato et al showed that the normalisation and fourth root pre-treatment, followed by the Modified Square Cosine Function was the most appropriate combination for intelligence purposes as it minimised false negatives. A few other comparison metrics were also evaluated to determine whether the Modified Square Cosine Function was still the optimal option. The comparison metrics evaluated from this project include two similarity measures, the Square Cosine Function and the Pearson Correlation. Additionally, three distance measures were also evaluated: the Euclideanm, Manhattan and Canberra distance.
So, how does this all tie in with prioritising the analytical techniques? Well, depending on the pretreatment and comparison metric selected, the separation between linked and unlinked populations will be different. Receiver operating characteristic curves are a common way to assess the overall separation between populations. A ROC curve is a plot of the true positive rate as a function of the false positive rate. So the closer to 1 the area under the curve is, the more efficient the comparison method. Here you can visualise what this population separation may look like. Choosing the wrong combination of pre-treatment and comparison metric may result in a large overlap between populations. However, with any data set, you will always observe some crossover between the two populations due to the presence of this error, scores need to be evaluated. In the deterministic approach, we try to determine the false positive and false negative routes associated at various threshold values, or comparison metric scores. It is usually up to the law enforcement agency to determine the final threshold value, as this will reflect the aims and resources. Once the threshold value is set, a new sample being tested against the database will have scores either falling above or below the threshold value, indicating a link or a lack of connection between it and respective specimens. As well as evaluating links between two specimens, we can group interlinked profiles through clustering algorithms. Hierarchical clustering is a common clustering algorithm used in illicit drug profiling. It typically joins nearby points into a cluster and then successively adds closer points to the nearest group. Subsequently, you end up with a dendogram, a sort of connectivity plot, which you can see on the screen. You can use this plot to decide after the fact how many cluster your data has. And this is done by cutting the dendogram at different heights.
To avoid any confusion, I'd like to bring you back to this slide. You'll notice now on the x-axis, we have the Modified Square Cosine function. With this function, a score closer to zero indicates examples are similar. While a score closer to 100 indicates that specimens are dissimilar. By plotting the distribution of scores and generating ROC curves, it was determined that the GC-MS profiles showed the greater separation between linked and unlinked populations. This is in comparison to the IRMS and chiral profiles. For this reason, the GC-MS data was used solely in subsequent analysis. Furthermore, the Modified Square Cosine function paired with normalisation and fourth root pre-treatment was deemed to be the optimal combination and was also used for the subsequent analysis.
The previous slide was a schematic of the separation. Here we can see the actual data. Due to the nature of this data set, there is quite a bit of overlap between the linked and unlinked populations. To make sure that this model can accurately differentiate between specimens which are linked and unlinked, we need to set an appropriate threshold value. In this case, the threshold value was based on a false positive rate of 2.5 percent, and this equates to a modified squared cosine score of approximately 30.
So once the threshold value has been set, we can use clustering analysis to create classes amongst the specimens to start to look at different trends and patterns in the data.
Some of the initial trends that I had a look at were generated through performing temporal analysis. By observing the evolution of chemical links over time, for instance, by comparing consecutively the map of links obtained trimester by trimester. Such visualisations are particularly useful to observe the emergence or extinction of criminal networks, as well as to allocate operational resources consequently. Indeed, investigative focus and efforts should primarily be targeted at clusters which are gaining importance across trimesters, with the aim of neutralising them as quickly as possible. Another way of visualising the movement of clusters of specimens over the years was through generating a six set Venn diagram. Interestingly, we can see that in the middle there were six clusters which were present throughout 2011 to 2016. This is much longer than the average cluster lifetime, which was determined to be about 2.2 years. By using temporal analysis, we can evaluate market dynamism. Interestingly, the effect of operations aimed at neutralising illicit drugs. Illicit drug selling groups could be evaluated using these indicators. For instance, an operation aimed to disrupt organised crime activities should theoretically lead to observe first and increase of the chemical links. Due to greater and more targeted attention. Then they should be followed by a decrease in the percentage of chemical links when these groups are effectively disrupted. Through conducting spatial analysis, it was determined that 51.4 percent of clusters connected seizures made in two or more jurisdictions. Furthermore, more than half of all seizures were made in two states, which is New South Wales and Western Australia. Therefore, it is not surprising to note that most of the clusters constitute specimens that were seized in New South Wales, Victoria and Western Australia. Out of the one hundred and twenty clusters observed, New South Wales seizures belong to 70 percent of them. Interestingly, 40 percent of those were exclusively composed of New South Wales seizures. All these trends serve as an indicator of the high methamphetamine market connectivity and especially the extent of trans jurisdictional activity. The results of studying this activity can guide decisions regarding collaboration or an increasing collaboration between relevant jurisdictions.
In addition to the temporal and spatial analysis performed, I also wanted to generate network plots between seizures and the respective clusters. Although there are many different ways to create network plots. I chose to use R to create a web application. This allowed me for more customised ability in the app compared to some of the other software currently available on the market. The idea is that all the relevant analysis will happen behind the scenes. So this includes a temporal and spatial analysis and an analyst will be able to observe in real time any trends or patterns surfacing in a particular drug market. On the screen, you could see a short example of how this app works.
In conclusion, it was possible to prioritise the available analytical techniques, where it was found that GC-MS, this was the most discriminative. Additionally, using organic impurities and clustering analysis, this study was able to infer the structure of the methamphetamine market in Australia. Furthermore, it was possible to infer when the market was the most structured and where. Finally, I would like to thank my supervisors and the industry partners for all of their help and input in this project. And I would also like to acknowledge that this work was supported by an Australian Research Council grant. Thank you all for listening and I hope you enjoyed the talk.
Thank you. Thank you for three speakers and I'd like to end the seminar. I'd like to thank all speakers and all of you who attended this seminar and I would like to invite everyone to close the session and to move back to the plenary session for the overall Q&A wrap up.
