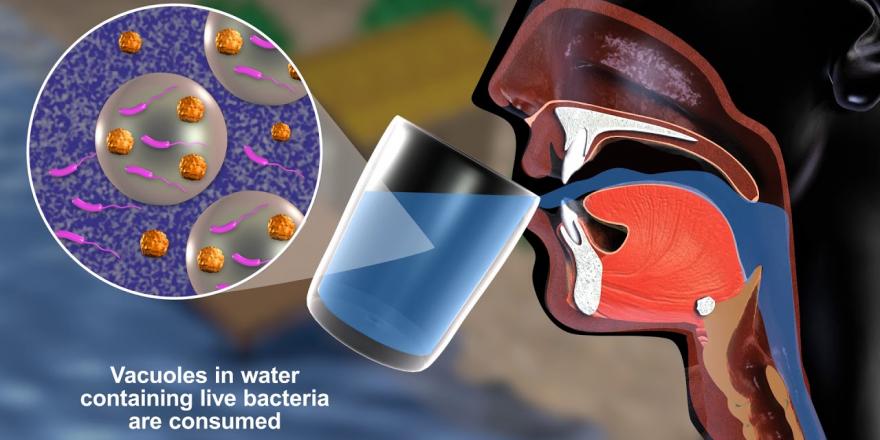
It started, as game-changing discoveries often do, with a simple observation. Back in 2012, a student in Associate Professor Diane McDougald’s Sydney lab, Parisa Noorian, noticed something weird: when she fed the bacteria Vibrio cholerae to protozoa – shape-shifting single-cell organisms that live in water and soil – they didn’t just eat lunch. Instead, the protozoa spat the bacteria out, undigested, as packages.
Why? Did they suspect the bacteria would make them sick? Or had the bacteria found a way to avoid being digested? What was going on?
In answering the question, McDougald and her team uncovered an important new story about the way virulence evolves in microbes, many of which present a significant risk to humans and their activities. The work has attracted the attention of microbiologists around the world and earned the small, nimble group of researchers from UTS the prestigious 2021 Australian Museum UNSW Eureka Prize for Scientific Research.
Over a painstaking decade, McDougald, colleague Dr Gustavo Espinoza-Vergara and their team at the UTS iThree Institute found that the cholera bacteria expelled by the protozoan host had, in fact, been turbocharged in the process. Protected inside the packages, known as expelled food vacuoles or EFVs for short, the bacteria were stronger, ten times more infective than they had been, impervious to antibiotics and to most attempts to kill them. The unexpected discovery has powerful implications for detecting dangerous bacteria, combatting microbial resistance to antibiotics, safeguarding drinking water, for farming and aquaculture sustainability and for predicting emerging diseases.
For McDougald, the result is an especially satisfying vindication of what she regards as an often-neglected approach; her team looked specifically to the environment, not the human body, for clues about the evolution of disease-causing bacteria.
The environmental lens
“As scientists, we study diseases from a human perspective: we’re always looking at how disease affects the human,” she says. “But actually, most of the bacteria that cause diseases are picked up from the environment. By ignoring their life – in water, in soil – we’re not learning how they can change and adapt to different environments. So, to look at them only after they infect us, I think, is a mistake.
“We can learn so much more by looking at the environment and how they interact with other organisms, which is what we’ve done. That’s the unique thing that we bring to this project.”
- Associate Professor Dianne McDougald
Bacteria have been on Earth far, far longer than humans but for the most part, we manage to live together: many bacteria do us no harm. But some evolve virulence and, in the process, become a danger. McDougald’s team was interested in understanding the factors that affect that evolution in its natural aquatic environment.
For their ground-breaking research, McDougald’s team focussed on Vibrio cholerae, the bacteria responsible for one of the most feared diseases and the death of millions. Despite the development of treatments and a strong understanding of disease biology in humans, cholera still infects more than one million people every year, often children in the least developed parts of the world. As McDougald notes, cholera remains a constant danger, especially after natural disasters or during war.
But dangerous as V. cholerae can be to humans exposed to contaminated food or water, it’s an opportunist, not well adapted to the high-acid environment of the human stomach. It takes many, many millions of free-living V. cholerae to infect a human but the team has established that far fewer ‘packaged’ bacteria – EFVs – can have a significant impact. The EFVs, as a vector, afford protection and create stronger and more infective bacteria, resistant to antibiotics and most disinfection measures.
Mysterious EFVs
“There’s something about being in the packages that makes them grow really fast, makes them hyper-effective, and super resistant. We don’t understand all of it yet,” McDougald explains. The role of EFVs explains one mystery that has baffled microbiologists – how V. cholerae remains in aquatic environments even between outbreaks when it is not detected by sampling.
One way to understand the implications of the UTS team’s discovery is to think of it applied to a city’s water storage. Authorities could have ways to detect V. cholerae in water, but without knowing whether the bacteria are free living or packaged in hyper-infectious EFVs, they may not understand the level of danger/threat until it is too late.
The initial research revealed that other opportunistic pathogens, also dangerous to humans, can survive in EFVs. McDougald says that bacteria including Salmonella, E. coli and Legionella, responsible for the dangerous pneumonia-like Legionnaires’ disease, are also likely to be rendered hyper-infectious by the journey through the vector.
“Legionella survives in air conditioner cooling towers and it’s 42 degrees – that's hot for a bacterium to survive. But we’ve taken Legionella in these vacuoles and put them in water at 45 degrees, and they're fine because they're so protected.”
The UTS team’s findings, published in the prestigious journal Nature Microbiology, have significant implications for everything from the health of farmed oysters, to understanding antibiotic resistance to possibly stamping out cholera for good.
“I think the takeaway for me about this work – about winning the Eureka Prize – is that it takes a long time to build a nice story,” says McDougald. “People shouldn’t get discouraged. I'm very aware that students get frustrated, and they go, ‘Well, it didn't work and we're not getting anywhere …’ But we are!"
Research team
-
iThree Institute
-
iThree Institute
-
iThree Institute
-
IThree Institute
-
iThree Institute
-
Dominic LeoIThree Institute
-
Jonah MooniThree Institute